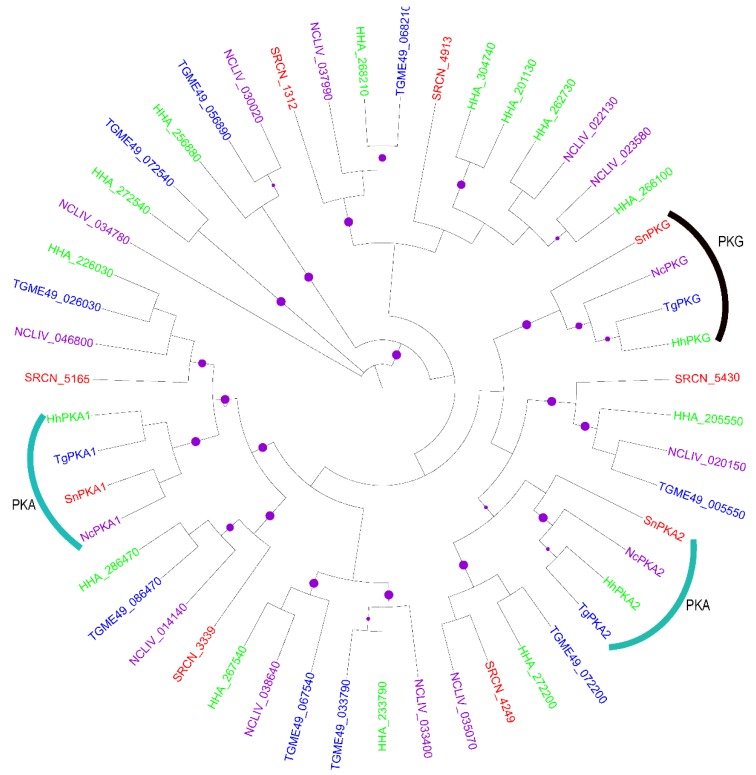
Figure 1.
Mid-point rooted maximum likelihood (ML) phylogenetic tree of apicomplexan AGCs. The terminal branches are color-coded for AGCs in the kinomes of Sarcocystis neurona (SRCN; red), Toxoplasma gondii, ME-49 strain (TGME49; blue), Hammondia hammondi (HHA; green) and Neospora caninum, Liverpool strain (NCLIV; purple). A solid purple circle on a branch indicates bootstrap support greater than 70. The phylogenetic tree was inferred from a multiple sequence alignment using PhyML with the Le and Gascuel (LG) amino acid substitution model and the gamma model of substitution rate heterogeneity. The tree image was rendered with iTOL.