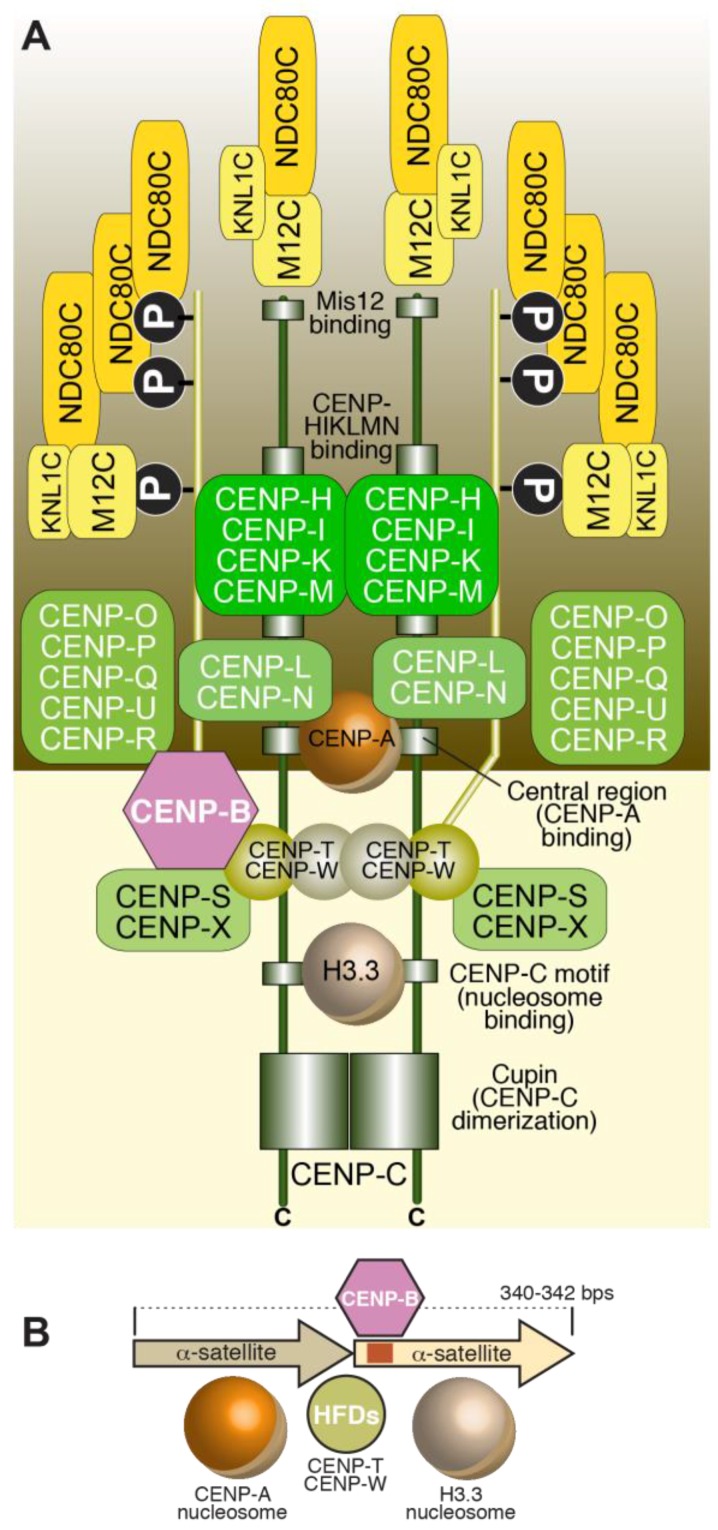
Figure 8.
A model for the assembly unit of the human kinetochore. The kinetochore assembly unit is depicted as being organized on a CENP-A-H3.3 dinucleosome. (A) A primary determinant of the stoichiometry of kinetochore subunits is the valency of the CENP-A nucleosome, which confers the potential to interact with two CCAN complexes, as shown in work of in vitro reconstitution of kinetochore assembly [110]. Because CENP-C and CENP-T each carries a full KMN network, and CENP-T additionally carries two NDC80 complexes, there are four MIS12 and KNL1 complexes per CENP-A nucleosome, and up to 8 NDC80 complexes. CENP-C has the potential to interact with two nucleosomes (see Figure 3), with one of them (the one bound to the CENP-C central region) is a CENP-A nucleosome permanently marked by interactions with stably bound CCAN subunits. As clarified in Figure 9, we speculate that the identity of the second nucleosome, which binds to the CENP-C motif, varies during the cell cycle, alternating between CENP-A and H3.3. The C-terminal dimerization domain of CENP-C might “seal” this design. During mitosis, the second nucleosome is an H3.3 nucleosome; (B) This speculative design is compatible with the existence of the tandem α-satellite structures already discussed in Figure 2. CENP-TW is proposed to bind in the inter-nucleosomal linker region, near the CENP-B box.