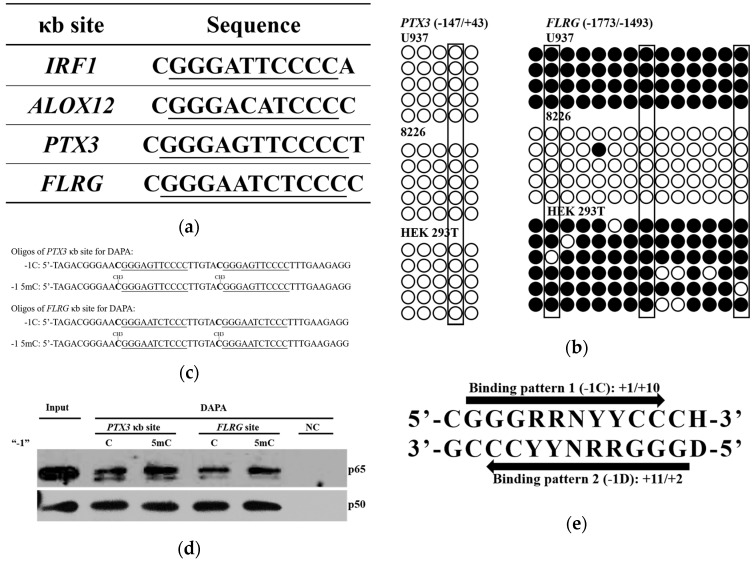
Figure 6.
The -1C methylation might not affect NF-κB binding when there are other binding possibilities. (a) Sequences of -1C κB sites with multiple binding patterns. The κB site sequences were underlined; (b) The methylation status of PTX3 and FLRG κB sites and their surrounding sequences, analyzed by BSP in U937, 8226 and HEK 293T cells. The CpGs at -1 position of κB sites are outlined. Methylated and non-methylated CpG sites are shown as solid or empty circles, respectively; (c) Sequences of the oligonucleotides with tandem PTX3 or FLRG κB sites used in DAPA. The κB site sequences were underlined, the nucleotides at -1 position were in bold; (d) Increased binding capacities of p65 and p50 with oligonucleotides bearing 5mC residues at -1C position of κB sites. DAPA were carried out using oligonucleotides containing κB sites of PTX3 or FLRG genes shown in Figure 6c and lysates of HEK 293T cells treated with TNFα (10 ng/mL) for 30 minutes. Oligonucleotide-bound NF-κB was detected by Western blot analysis using antibodies against p65 or p50, respectively. Data shown are the results of one of three experiments. NC, negative control oligonucleotide. Input, 2% of the whole cell lysate used in DAPA; (e) Possible κB sites in PTX3 and FLRG. D = A, T, G; H = T, A, C.