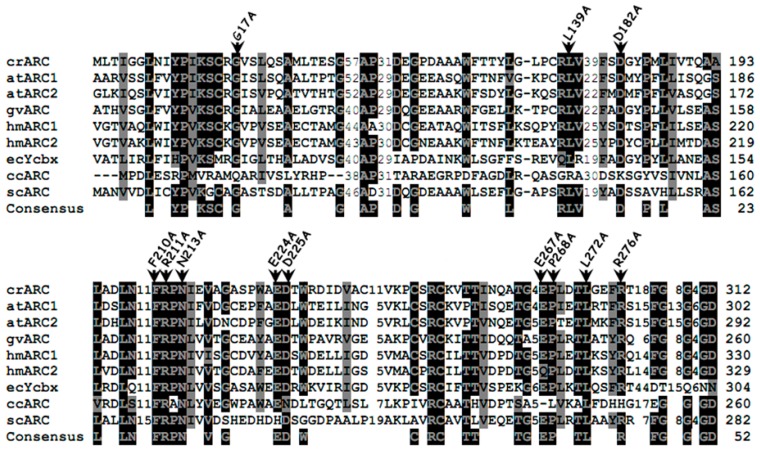
Figure 4.
Multiple-sequence alignment of different mARC proteins. The sequences and accession numbers (shown into parentheses; GenPept accession numbers begin with XP and NP; others are proteins deduced from GenBank sequences) are as follows: crARC, Chlamydomonas reinhardtii mARC (XP_001694549); atARC1, Arabidopsis thaliana mARC1 (NP_174376); atARC2, Arabidopsis thaliana mARC2 (NP_199285); gvARC Gloeobacter violaceus PCC 7421 mARC (NP_926027); hmARC1, human mARC1 (Homo sapiens) (NP_073583); hmARC2, human mARC2 (NP_060368); YcbX, Escherichia coli YcbX (NP_415467); ccARC, Caulobacter crescentus mARC (AAK22857); and scARC, Streptomyces coelicolor A3ARC (CAC04053). The positions of residues mutated to alanine in crARC are indicated by the black arrowheads. Highly conserved amino acids are shown on a black background, and moderately conserved amino acids are shown on a gray background. The consensus sequences have been calculated with a threshold of 75% with the BioEdit v.7.0.9 program (Reprinted with permission from [17]).