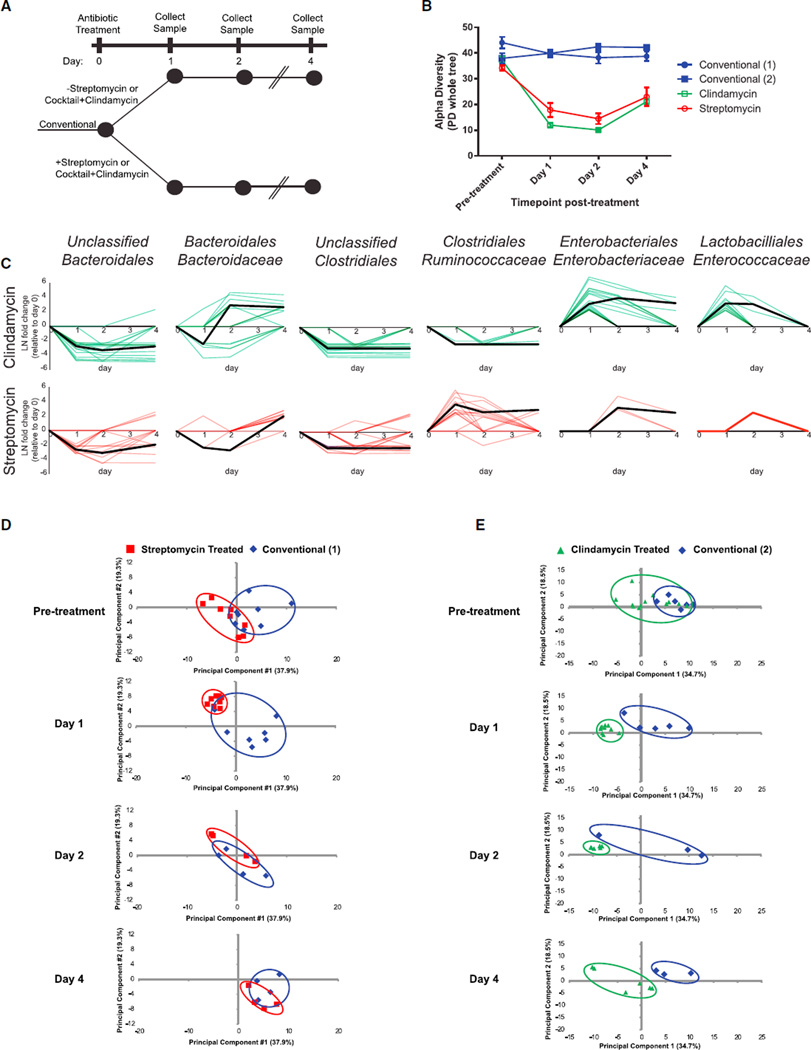
Figure 1. Antibiotic Treatment Elicits Distinct Effects on the Host and Microbiota.
(A) Conventional mice were dosed with one of two antibiotic treatments: streptomycin or a 2-day course of an antibiotic cocktail followed by a single dose of clindamycin. Antibiotic- and vehicle-treated mice were followed for 4 days, and stool samples were collected for 16S rRNA analysis and host-centric proteomics.
(B) Alpha diversity at each time point (mean ± SEM) measured by the PD_whole_tree metric from 16S rRNA sequencing data. Conventional (1) refers to the control mice from the streptomycin experiment, whereas Conventional (2) refers to the control mice from the clindamycin treatment.
(C) OTUs that are significantly affected by antibiotic treatment, grouped by microbial family. Briefly, DESeq was used to identify significantly changing OTUs and their respective fold changes (natural log, LN) when comparing a particular time point with the same mice recorded at day 0. For each comparison, each significantly changed OTU at any time point was plotted as a colored line individual point. Zero values are plotted for insignificantly or unchanged OTUs and time points. Black lines represent the median value of OTU fold changes that significantly deviated from day-0 levels.
(D and E) Principal-component analysis was conducted on all 1,425 host proteins identified across all samples in the (D) streptomycin and (E) clindamycin experiments and plotted by time point. Diminishing numbers of mice exist because mice were put into different arms of the general experimental paradigm (Figure S1).