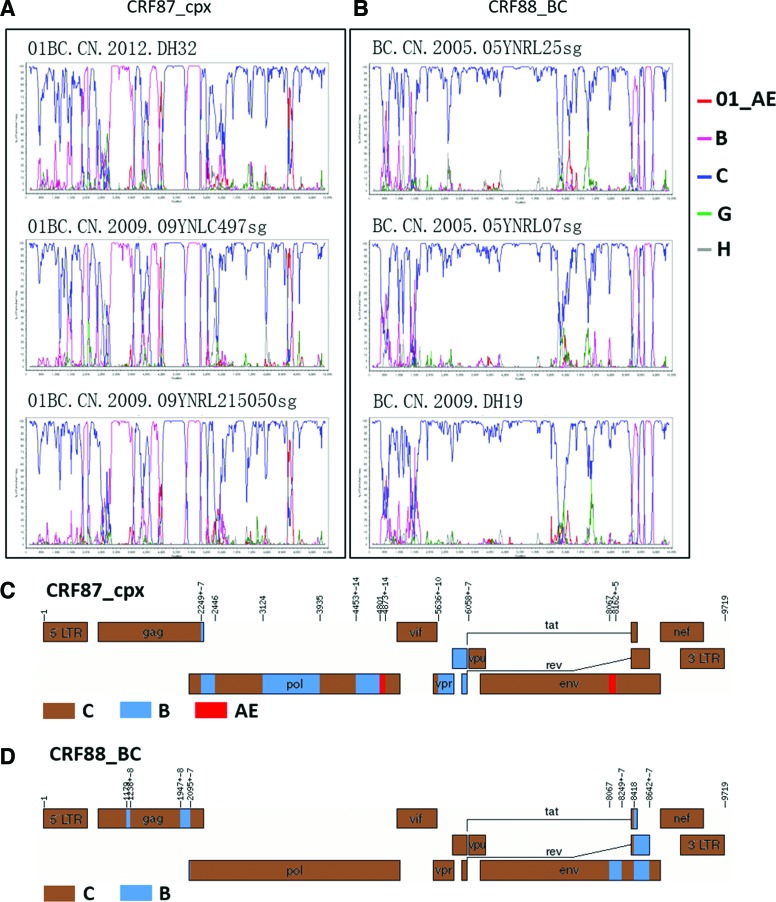
FIG. 2.
Two new CRFs identified in this study. (A) and (B) bootscan plots of three representative strains of CRF87_cpx and CRF88_BC, respectively. (C) and (D) Mosaic maps of CRF87_cpx and CRF88_BC, respectively. The parameters of the SimPlot bootscan analysis were window size, 300 bps; step size, 20bps; and GapStrip on. The schematic maps were generated by Recombinant HIV-1 Drawing Tool available at the LANL web site. Breakpoint positions were numbered according to HXB2. Color images available online at www.liebertpub.com/aid