Abstract
Low-Grade Gliomas can vary widely in disease course and therefore patient outcome. While current characterization relies on both histological and molecular analysis of tissue resected during surgery, there remains high variability within glioma subtypes in terms of response to treatment and outcome. In this study we hypothesized that parameters obtained from Magnetic Resonance (MR) data would be associated with progression-free survival (PFS) for patients with recurrent low-grade glioma. The values considered were derived from the analysis of anatomic imaging, diffusion weighted imaging, and 1H Magnetic Resonance Spectroscopic Imaging (MRSI) data. Metrics obtained from diffusion and spectroscopic imaging presented strong prognostic capability within the entire population as well as when restricted to astrocytomas, but demonstrated more limited efficacy in the oligodendrogliomas. The results indicate that multi-parametric imaging data may be applied as a non-invasive means of assessing prognosis and may contribute to developing personalized treatment plans for patients with recurrent low-grade glioma.
Keywords: Glioma, MRI, Diffusion, Progression-free Survival
Introduction
Infiltrating gliomas have a wide range of clinical outcomes [1–2]. Patients diagnosed with a low-grade glioma (LGG) may survive for decades, highlighting the need to balance treatment and quality of life for each patient. Defining the criteria for deciding when to treat and which therapies to apply remains challenging. The current standard-of-care for patients newly diagnosed with a low-grade glioma consists of surgical resection followed by a choice of active surveillance, chemotherapy, or radiation therapy [3]. Histological sub-type can inform these decisions. These subtypes have been defined as astrocytoma, and oligodendroglioma, and mixed oligoastrocytoma. As molecular characteristics based upon isocitrate dehydrogenase (IDH) mutation and 1p19q deletion status can provide important prognostic information [4–5], diffuse gliomas are now classified using both histologic and molecular characteristics [6]. Using these markers, the new WHO classification no longer has the mixed glioma category.
Identifying biomarkers that are able to non-invasively assess tumor aggressiveness and stratify patients into distinct subgroups with more uniform clinical outcome is critical for making treatment decisions, especially at the time of recurrence. Diffusion weighted imaging (DWI) provides information about tissue architecture and cellularity [7]. The Apparent Diffusion Coefficient (ADC) is used as a measure of cellularity [8–11], and low ADC is associated with shorter progression free and overall survival in LGGs [11–13]. ADC may distinguish between different histological sub-types of LGG [14], and lesions that have transformed to a higher grade and are likely to fail treatment early [15–16]. Fractional Anisotropy (FA) is a measure of the degree of directionality in diffusion [7]. FA has been associated with histology and patient outcome [17–18].
1H spectroscopic imaging (MRSI) provides spatially localized metabolic and chemical information such as levels of choline-containing compounds (Cho), creatine (Cr), N-acetylaspartate (NAA), lactate (Lac), and lipid (Lip) [19]. The Cho signal reflects changes in membrane synthesis and turnover associated with cell proliferation and remodeling. The Cr peak includes both creatine and phosphocreatine and is indicative of cellular bioenergetic processes. NAA is a marker of normal brain tissue associated with the presence of actively functioning neurons. Lac is an end product of anaerobic metabolism and reflects ischemia and/or hypoxia. Lip reflects the presence of necrosis. Levels of these metabolites have been linked to shorter progression free and overall survival in both GBMs [20–22] and LGGs [23].
Recent studies using these advanced, multi-parametric imaging methods to target image guided tissue sampling in patients with recurrent LGG have highlighted characteristics that are able to predict malignant transformation [17]. The next step is to determine whether similar criteria can be applied to non-invasively assess prognosis and indicate if/when a change in treatment strategy may be beneficial. The current study addresses this challenge in patients with recurrent LGGs by testing the hypothesis that MR anatomic, diffusion and spectroscopic parameters obtained at the pre-surgery examination provide important prognostic information. The imaging parameters considered included summary metrics derived from histogram analysis of anatomic T1-contrast and Fast Spin Echo (FSE) image intensities, ADC, FA, Cho to NAA index (CNI) and normalized levels of Lac. The associations of imaging parameters with PFS were evaluated for astrocytoma and oligodendroglioma subgroups. Parameters that were identified as relevant for defining prognosis were examined to test their relationship to histological parameters derived from image guided tissue samples.
Materials and Methods
The participants in this IRB-approved study comprised adult patients with an original diagnosis of a grade II glioma who were presenting for surgery with suspected recurrence. All subjects had a biopsy or surgical resection at original diagnosis. For the majority of patients, there had been one or more prior recurrences (defined by imaging criteria), but in all cases the current surgery was the first surgery for recurrence. A total of 122 patients received MR examinations immediately prior to surgery. Image-guided tissue samples were obtained during resection and used to evaluate histological parameters and molecular characteristics. All patients gave informed consent to participate in the study.
MR imaging and spectroscopy
Anatomical, diffusion weighted, and spectroscopic imaging were acquired using either 1.5 T or 3 T GE scanners with an eight channel phased-array headcoil (MRI Devices). Anatomical imaging included T2-weighted Fluid Attenuated Inversion Recovery (FLAIR), Fast Spin Echo (FSE), and T1-weighted images acquired pre- and post-gadolinium injection. Diffusion weighted imaging (DWI) was acquired axially with six gradient directions and a voxel size of 1.7mm x 1.7mm x 3mm (TR/TE = 1000/108ms and b=1000 s/mm2). 3D Lactate-edited proton MR spectroscopic imaging (1H MRSI) was acquired with (CHESS) water suppression, point-resolved spectroscopic selection (PRESS) localization, and very selective saturation (VSS) pulses for outer volume suppression with TE/TR = 144/1104ms, matrix size = 16x16x16 and a nominal voxel size of 1x1x1 cm3. A flyback echo-planar k-space trajectory was used to sample data in the SI direction in order to decrease the total acquisition time [24].
Custom software was used to calculate parameters from DWI and 1H MRSI. A previously published algorithm [7] was used to calculate ADC and FA maps. All anatomic images and diffusion maps were aligned to the T1 post-contrast image using FSL’s Linear Image Registration Tool (http://fsl.fmrib.ox.ac.uk). Spectroscopic data were reconstructed in each channel and combined in order to quantify the relevant metabolites choline (Cho), N-acetyl-aspartate (NAA), and lactate (Lac). The choline-to-NAA index (CNI) was calculated for spectroscopic acquisitions using a published algorithm [25].
Manually defined regions of interest (ROIs) were created to delineate normal appearing brain tissue (NABT), the T2-hyperintensity (T2ALL) lesion on the FLAIR images, and the enhancing lesion (CEL) on the post-contrast T1 images. A T1-subtraction image was generated to delineate the region of interest (T1s) corresponding to enhancement within the T2ALL. Volumes of each of these ROIs were recorded. The values of within these ROIs from FSE images, ADC and FA maps were normalized to the mode of intensities in NABT. Percentiles of normalized intensity (10th, 50th, 90th) were calculated for histograms from within the T2ALL lesion. The fraction of voxels within the T2ALL lesion with normalized ADC (nADC) <1.5 was recorded. The metabolic lesion was defined as the region with CNI>2 and its volume (CNI2) was recorded.
Levels of Lac were normalized (nLac) by the median level of NAA from voxels within the selected volume but outside of the CNI2 region. The integrated sums of CNI (sumCNI) and nLac (sumLac) values from voxels within the CNI2 that were within or partially within the T2ALL were used as measures of metabolic abnormality.
Image-Guided Tissue samples
Image-guided tissue targets were chosen based on surgically accessible regions of abnormally decreased ADC, increased cerebral blood flow, and elevated CNI. Targeted tissue samples were excised based on neurosurgical guidance using intra-operative navigation. 3D screenshots of the navigation system recorded the exact coordinates of each tissue sample. Tissue samples were fixed in 10% zinc formalin and dehydrated by graded ethanols before being embedded in Paraplast Plus wax (McCormick Scientific) using standard techniques for tissue processing and immunohistochemistry. We did not excise any tissue samples from normal brain.
Histopathology and Molecular Analysis
Tissue samples were reviewed and scored by a board-certified neuropathologist. Mouse monoclonal anti-IDH1R132H (DIA H09) antibody was used to screen samples for the presence of IDH1 mutation. This was performed in Tris-EDTA buffer at pH 8. Following antigen retrieval, sections were treated with 3% methanol-hydrogen peroxide for 16 min at 22ºC. Tumors were classified as IDH1 mutant or wildtype based on immunostaining. IDH wildtype status was not confirmed with IDH1 and IDH2 codon sequencing, so some of these tumors may harbor rarer IDH1 or IDH2 mutations. All immunohistochemistry assays were performed on the Ventana Medical Systems Benchmark XT.
Fluorescence in situ hybridization (FISH) was performed utilizing previously published methods (PMID 25165197) and commercially available paired probe sets to assess 1p19q deletion status using loci-specific identifiers (LSIs; Abbott Laboratories, North Chicago, IL). In brief, one probe set used LSI 1p36 with a 1q25 LSI reference for 1q. The other probe set used a 19q13 LSI with 19p13 reference probe for 19p. Formalin-fixed, paraffin-embedded tissue on two glass slides were de-paraffinized and then treated with pepsin. The DNA was then heat-denatured and subsequently allowed to hybridize with the two probe sets (one per slide). The slides were then washed and counterstained with DAPI (4,6 diamidino-2-phenylidole). Enumeration of 1p, 1q, 19p, and 19q signals was conducted with fluorescence microscopy using appropriate excitation and emission filters.
Assays for Ki-67 (to assess proliferation) and CA9 (to assess hypoxia) were performed on most samples obtained at surgery. Slides stained with anti Ki-67 (30–9) rabbit monoclonal (Ventana Medical Systems) at 2μg/ml for 23 min at 37ºC were scored for Ki-67 labelling index [(Ki-67-positive nuclei per total tumor cells counted per 200x field) x 100%] based on the evaluation of at least three fields and >1000 cells. Total cellularity was determined as an average number of cells per 200x field.
The degree of hypoxia was qualitatively measured using IHC stained sections for CA9 using a four-tier ordinal scale of immunoreactivity (0, no immunoreactivity; 1, <10% tumor cells immunopositive; 2, 10–25% tumor cells positive; 3, >25% tumor cells positive) within three high power fields at a magnification of 200x.
Clinical Parameters and Characterization
Tumor grade and subtype were determined by a board-certified neuropathologist. Sub-type classification was based on histology and molecular characteristics. LGG gliomas with astrocytoma or oligodendroglioma histology were classified accordingly, while gliomas with oligoastrocytoma histology were classified as oligodendrogliomas if they were 1p19q codeleted, and as astrocytomas if they were 1p19q intact. These oligodendroglioma and astrocytoma subtypes were further stratified as IDH mutant or IDH wildtype when IDH IHC status was known. Because 1p19q status was not available for all patients, and IDH wildtype status was not confirmed with IDH1 and IDH2 codon sequencing, immunonegative and/or 1p19q-unknown cases were best classified under the new 2016 WHO structure as astrocytoma NOS and oligodendroglioma NOS. However, we also report the results of the subgroups of astrocytoma, IDH-mutant and oligodendroglioma, IDH-mutant and 1p/19q-codeleted when known. We have included patients whose diagnosis at their first surgery was grade II glioma, but who were grade IV at this recurrent surgery. These patients were further stratified as IDH mutant or IDH wildtype based on IHC. These grade IV patients are included in the astrocytoma group because of their predominantly astrocytic composition.
Tumor progression was determined based upon RANO criteria using standard anatomic images [26]. The PFS was measured from the time of this surgery at recurrence until the subsequent progression or last known contact at UCSF. At the time of analysis, median overall survival had not been reached, so the relationship of imaging parameters to OS was not assessed. The extent of resection (EOR) was defined as gross-total or subtotal based upon standard radiological criteria applied to the clinical MR scan that took place within 24 hours of surgery.
Analysis of Imaging Parameters
To test for group differences in PFS between the grades at recurrence (II, III, and IV), histology subgroups, extent of resection (EOR), and subsequent chemotherapy or radiation treatment, Kaplan-Meier curves were plotted for patient subgroups and the significance of pairwise differences was judged using either a Log-rank test or, if the assumptions of the Log-rank test were violated, a Tarone-Ware test. In order to further assess the significance of grade differences in the astrocytomas, grade was used as sole variable in a Cox proportional hazards model. Additional comparisons within subgroups of patients that had IDH mutations were also performed. The small number of progression events observed for IDH wildtype patients precluded an analysis within that subgroup.
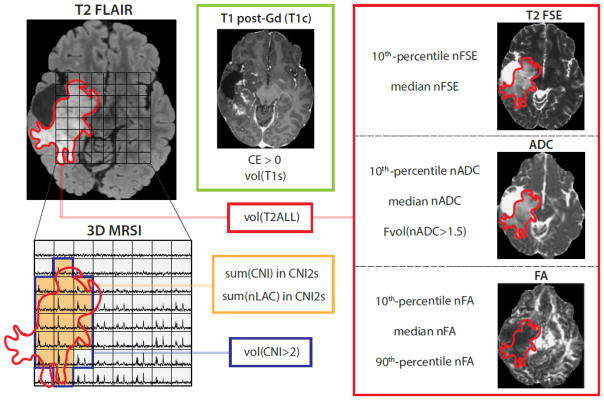
The 14 multi-modality imaging parameters evaluated in the analysis are represented in Figure 1. To test associations of imaging parameters with PFS, each parameter was modeled (controlling for age, EOR, and subsequent treatment) in a Cox proportional-hazards model. Treatment after surgery was coded as the presence or absence of RT, and the presence or absence of chemotherapy. Significance was assessed for individual parameters with a Bonferroni adjustment (p < .05/14 = .0036). This Cox regression analysis was also performed for each parameter within each histology subgroup and with subjects who had IDH mutation. Wilcoxon rank-sum tests were used to assess differences in imaging parameters between subgroups.
Figure 1.
Quantification of MR parameters used in the present analysis. nADC, FA, and FSE intensity percentiles are calculated within the T2 Flair hyperintensity (T2ALL) ROI. In the MRSI image, the blue line delineates voxels with CNI>2, while the green-shaded region (volCNI2) identifies voxels where CNI>2 voxels overlap or are tangent to T2ALL (CNI2s). The integrated sums of CNI (sumCNI) and nLac (sumLac) values from voxels within the CNI2 that were intersecting or at least tangent to the T2ALL were used as measures of abnormal metabolic features of the tumor.
To compare imaging with histopathology, 5mm spherical ROIs were generated at tissue sample locations and the median imaging value determined within the ROI. Correlation between these median values and histopathological parameters (Ki-67, CA9) were determined by iteratively randomly choosing one biopsy per patient and performing a Kendall Tau correlation test on the values from those randomly selected biopsies, as well as a single Kendall Tau test that included all samples from all patients.
In order to enhance the utility of the links between imaging and PFS found in our analysis, we employed a colormapping scheme that exploits the differences in population-wide histograms to create a function that converts a pixel intensity into an RGB vector. The histograms of nADC map pixel intensity values come from normal tissue, lesions with high median intensity in the T2ALL, and those with low median intensity in the T2ALL. Each pixel in the image is colored by an RGB vector whose components are the heights (relative to respective maxima) of each histogram.
Results
Patient characteristics
Nine of the 122 patients who were recruited to the study were excluded due to a diagnosis inconsistent with diffuse glioma, and two additional patients were immediately lost to clinical follow-up after resection. This left 111 patients whose records were available for PFS analysis (54 females and 57 males). The median time from initial to current surgery was 2261 days (6.2 years). Forty-one percent of patients remained grade II, while 43% had upgraded to grade III and 15% had upgraded to grade IV. There were 18 patients with grade II astrocytoma (11 IDH mutant), 33 subjects with grade III astrocytoma (23 IDH mutant), 17 with grade IV astrocytoma (11 IDH mutant), 27 with grade II oligodendroglioma (20 IDH mutant), and 16 with grade III oligodendroglioma (all IDH mutant). Prior to the current surgery, 24% of the patients received radiation therapy, 41% received temozolomide (TMZ) and 7% received other experimental agents. Following the current surgery, 78% received chemotherapy and 60% received irradiation. Subsequent treatment was unknown for two patients.
Seventy-three percent of patients progressed (median PFS = 737 days). Among grade II patients, 70% progressed (median PFS = 847 days). Among grade III patients, 67% percent progressed (median PFS = 894 days). Among grade IV patients, all progressed (median PFS = 329 days). Seventy-two percent of the astrocytoma patients progressed (median PFS = 642 days). Seventy-three percent of the oligodendroglioma patients progressed (median PFS = 806 days).
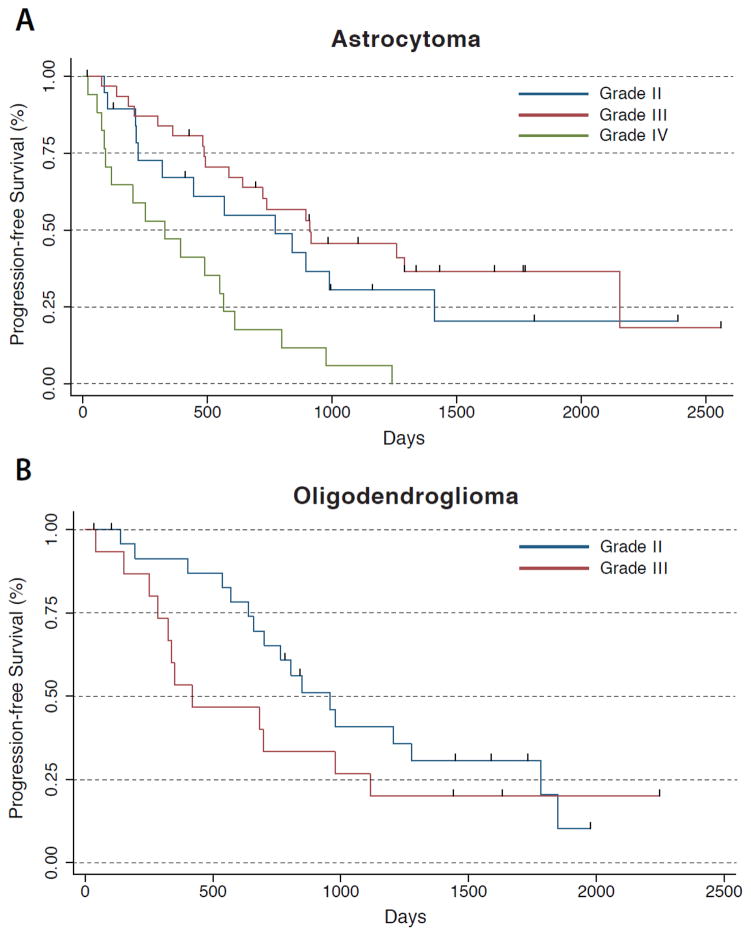
Kaplan-Meier curves of the PFS for all LGGs grouped by subtype and grade are shown in Figure 2. LGGs that recurred as grade IV had significantly shorter PFS than astrocytomas that recurred as grade II (P<0.05) or grade III (P<0.001). Within astrocytomas with IDH mutations, only grade III and grade IV were significantly different (P<0.01). There was no significant difference in PFS between grade II and grade III astrocytomas. There were no significant differences between grade II and grade III oligodendrogliomas, either for the overall subgroup or within IDH mutants. There were no significant differences between oligodendrogliomas and astrocytomas. LGGs with subtotal resections had significantly shorter PFS than LGGs with gross total resections (P<.05). LGGs treated with subsequent chemotherapy had significantly longer PFS than LGGs not treated with subsequent chemotherapy (P<.0001).
Figure 2.
(a) Among astrocytoma patients, patients recurring as grade IV have significantly worse PFS outcome than those recurring as grade II or III (who did not differ significantly). (b) Among oligodendroglioma patients, there is no significant difference in PFS outcome by grade.
Table 1 provides a summary of imaging parameter values and the associations between imaging parameters and PFS (controlling for age, EOR, and subsequent treatment). Across all patients, the volumes of the T2ALL and T1s ROIs, and the median and 10% FSE signal intensity were significantly associated with PFS. Within the DWI parameters, the Fvol(nADC>1.5) (the fraction of T2ALL with nADC > 1.5), median and 10% nADC, and median, 10% and 90% nFA were significantly associated with PFS. nADC values were lower and nFA values were higher in patients with poorer PFS. Within the MRSI parameters, sum(CNI) and sum(nLac) were significantly associated with PFS. CNI and Lac were higher in patients with poorer PFS. All of these MR parameters except 10% nADC and sum(nLac) were also significantly predictive of PFS in the astrocytomas, while only the nFA parameters were significantly predictive of PFS in the oligodendrogliomas. Within the IDH mutant astrocytomas, T2ALL vol, Fvol(nADC>1.5), 10%, median, and 90% nFA, sum(CNI) and the CNI>2 volume were significantly associated with PFS. Within the IDH mutant oligodendrogliomas, only 10% nFA was significantly associated with PFS.
Table 1.
Median imaging parameter values, ranges, and Cox proportional hazards model imaging coefficients in the overall patient population and within each subgroup (controlling for age, extent of resection, and subsequent treatment). p values falling below the Bonferroni alpha correction of .0036 are bolded.
| Imaging parameter | Media n (range) | All patients | Astrocytoma | Oligodendroglioma | ||||
|---|---|---|---|---|---|---|---|---|
| N | p value | N | p value | N | p value | |||
| vol(T2ALL) | 28 cc (1 – 540) | 108 | 0.003* | 67 | 0.005 | 38 | n.s. | |
| CE > 0 (Y/N) | 57 N, 53 Y | 108 | n.s. | 67 | n.s. | 38 | n.s. | |
| vol(T1s) | 1 cc (0 – 146) | 95 | 0.024 | 59 | 0.049 | 33 | n.s. | |
| nFSE | 10 % | 1.4 (0–2.34) | 107 | 0.006 | 66 | 0.035 | 38 | n.s. |
| Median | 2.19 (1.11 – 4.3) | 106 | 0.014 | 66 | 0.018 | 37 | n.s. | |
| Fvol(nADC>1.5) | .31 (.04 – .91) | 103 | <0.001* | 64 | <0.001* | 36 | n.s. | |
| nADC | 10 % | 1.22 (.29 – 1.78) | 103 | 0.006 | 64 | n.s. | 36 | n.s. |
| Median | 1.66 (1.08 – 2.6) | 103 | 0.006 | 64 | 0.009 | 36 | n.s. | |
| nFA | 10 % | .32 (.17 – .61) | 101 | <0.001* | 63 | <0.001* | 35 | 0.011 |
| Median | .83 (.50 – 1.61) | 101 | <0.001* | 63 | <0.001* | 35 | 0.024 | |
| 90% | 1.54 (1.10 – 2.72) | 101 | <0.001* | 63 | 0.004 | 35 | 0.027 | |
| vol(CNI2) | 21 cc (.50 – 139) | 81 | n.s. | 47 | <0.001* | 31 | n.s. | |
| sum(CNI) in CNI2s | 62 (.16 – 1020) | 78 | .022 | 45 | 0.004 | 30 | n.s. | |
| sum(nLac) in CNI2s | 2.27 (0 – 85) | 78 | .040 | 45 | n.s. | 30 | n.s. | |
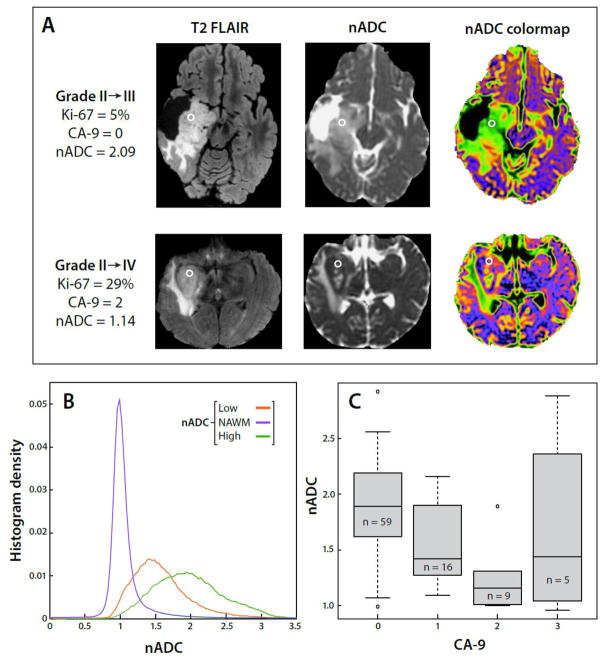
Given that multiple DWI parameters were associated with outcome for astrocytoma patients, we assessed the relationship between median nADC values from locations at which 91 tissue samples had been obtained from 52 astrocytoma patients and histological markers from those tissue samples. The correlation of median nADC values with Ki-67 was significant (P=0.022, accounting for the multiplicity of samples per subject). Median nADC values were also significantly related to CA9 scores (p=.028) (Figure 3). In both cases, lower nADC values were related to more malignant tissue characteristics (Figure 4).
Figure 3.
(a) Examples of a Grade II→III patient and Grade II→IV patient demonstrating differing nADC and levels of Ki-67. The circled regions represent tissue sample locations. (b) Average nADC histograms for the patient population, consisting of normal-appearing white matter (NAWM) and low and high nADC regions, which were used to produce the nADC colormaps (c) A boxplot comparison of nADC and extent of hypoxia, as denoted by increasing CA9 score, within astrocytoma patients. Values over one inter-quartile range beyond the 25th or 75th percentiles were plotted as outliers.
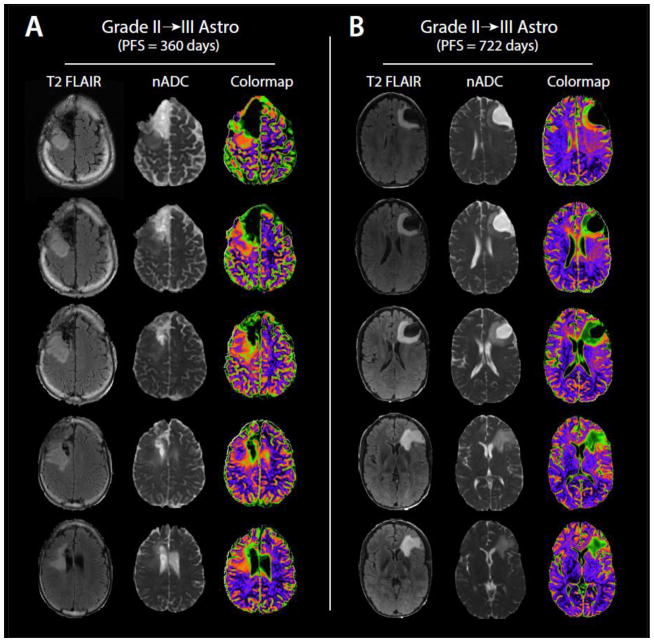
Figure 4.
(a) Example of T2 FLAIR, nADC, and nADC colormap images from the exam of a 26 year old Grade II→III patient with a PFS of 360 days. (b) Example of Flair, nADC, and nADC colormap images from the exam of a 28 year old Grade II→III patient with improved PFS outcome of 722 days.
Discussion
In this study we hypothesized that Magnetic Resonance (MR) imaging and spectroscopic parameters would be associated with progression-free survival for patients with recurrent low-grade glioma. These parameters were derived from the analysis of anatomic imaging, diffusion weighted imaging, and 1H Magnetic Resonance Spectroscopic Imaging (MRSI) data. Diffusion and spectroscopic imaging were predictive of PFS within the entire population, and within astrocytoma and oligodendroglioma subgroups. These results suggest that imaging provides a non-invasive means of assessing prognosis that may be used in developing personalized treatment plans for patients with recurrent low-grade glioma.
One of the major limitations of analyses of progression-free and overall survival is that these analyses must be delayed until at least half of the patient population has experienced progression or death. We are continuing to clinically follow the patients in this study, and will directly assess the degree to which these imaging data predict OS when median OS for is reached. Future studies will also benefit from emerging advanced imaging techniques such as spectroscopic detection of 2-hydroxyglutarate [29] and 13C spectroscopy [30].
Ultimately, the ability to identify lesions and target biopsies to regions that are associated with poor outcome could provide clinicians with an enhanced ability to diagnose and treat this challenging disease. Longitudinal observation of imaging parameters may allow us to track residual tumor regions post-surgery, identify treatment response, and detect early signs of progression. Ongoing projects at our institution are building on these methodologies and investigating their capability in patient populations of newly diagnosed LGG, as well as in combination with genomic and epigenomic analyses of the tumors. With the addition of new prognostic imaging biomarkers to the current paradigm, non-invasive quantitative imaging could play an increasingly important role in improving the clinical management and quality of life for these patients.
Acknowledgments
We thank the staff of the UCSF Margaret Hart Surbeck Laboratory and the UCSF Brain Tumor SPORE Tissue Bank. This research was funded by the UCSF Brain Tumor SPORE (P50 CA097257)
Footnotes
Ethical approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
References
- 1.Shaw EG, Wisoff JH. Prospective clinical trials of intracranial low-grade glioma in adults and children. Neuro-Oncology. 2003;5(3):153–160. doi: 10.1215/S1152-8517-02-00060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaeckle KA, Decker PA, Ballman KV, Flynn PJ, et al. Transformation of low grade glioma and correlation with outcome: an NCCTG database analysis. Journal of Neuro-Oncology. 2011;104(1):253–259. doi: 10.1007/s11060-010-0476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grier JT, Batchelor T. Low-Grade Gliomas in Adults. The Oncologist. 2006;11(6):681–693. doi: 10.1634/theoncologist.11-6-681. [DOI] [PubMed] [Google Scholar]
- 4.Wang H-Y, Tang K, Liang T-Y, Zhang W-Z, et al. The Comparison of Clinical and Biological Characteristics between IDH1 and IDH2 Mutations in Gliomas. Journal of Experimental & Clinical Cancer Research : CR. 2016;35:86. doi: 10.1186/s13046-016-0362-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leeper HE, Caron AA, Decker PA, Jenkins RB, Lachance DH, Giannini C. IDH mutation, 1p19q codeletion and ATRX loss in WHO grade II gliomas. Oncotarget. 2015;6(30):30295–30305. doi: 10.18632/oncotarget.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 7.Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. Journal of Magnetic Resonance. 1996;111(3):209–19. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- 8.Wen Q, Jalilian L, Lupo JM, et al. Comparison of ADC metrics and their association with outcome for patients with newly diagnosed glioblastoma being treated with radiation therapy, temozolomide, erlotinib and bevacizumab. Journal of Neuro-Oncology. 2015;121:331–339. doi: 10.1007/s11060-014-1636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilario A, Sepulveda JM, Perez-Nuñez A, et al. A Prognostic Model Based on Preoperative MRI Predicts Overall Survival in Patients with Diffuse Gliomas. American Journal of Neuroradiology. 2014;35:1096–1102. doi: 10.3174/ajnr.A3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romano A, Calabria LF, Tavanti F, Minniti G, Rossi-Espagnet MC, Coppola V, Pugliese S, Guida D, Francione G, et al. Apparent diffusion coefficient obtained by magnetic resonance imaging as a prognostic marker in glioblastomas: correlation with MGMT promoter methylation status. European Radiology. 2013;23:513–520. doi: 10.1007/s00330-012-2601-4. [DOI] [PubMed] [Google Scholar]
- 11.Zulfiqar M, Yousem D, Lai H. ADC Values and Prognosis of Malignant Astrocytomas: Does Lower ADC Predict Worse Prognosis Independent of Grade of Tumor? – A Meta-Analysis. American Journal of Roentgenology. 2013;200:624–629. doi: 10.2214/AJR.12.8679. [DOI] [PubMed] [Google Scholar]
- 12.Cuccarini V, Erbetta A, Farinotti M, et al. Advanced MRI may complement histological diagnosis of lower grade gliomas and help in predicting survival. Journal of Neuro-Oncology. 2016;126(2):279–288. doi: 10.1007/s11060-015-1960-5. [DOI] [PubMed] [Google Scholar]
- 13.Caseiras GB, Chheang S, Babb J, et al. Relative cerebral blood volume measurements of low-grade gliomas predict patient outcome in a multi-institution setting. European Journal of Radiology. 2010;73(2):215–220. doi: 10.1016/j.ejrad.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Khayal IS, VandenBerg SR, Smith KJ, et al. MRI apparent diffusion coefficient reflects histopathologic subtype, axonal disruption, and tumor fraction in diffuse-type grade II gliomas. Neuro-Oncology. 2011;13(11):1192–1201. doi: 10.1093/neuonc/nor122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta RK, Cloughesy TF, Sinha U, Garakian J, Lazareff J, Rubino G, Rubino L, Becker DP, Vinters HV, Alger JR. Relationships between choline magnetic resonance spectroscopy, apparent diffusion coefficient and quantitative histopathology in human glioma. J Neurooncol. 2000;50:215–26. doi: 10.1023/a:1006431120031. [DOI] [PubMed] [Google Scholar]
- 16.Brunberg JA, Chenevert TL, McKeever PE, et al. In vivo MR determination of water diffusion coefficients and diffusion anisotropy: correlation with structural alteration in gliomas of the cerebral hemispheres. Am J Neuroradiol. 1995;16:361–371. [PMC free article] [PubMed] [Google Scholar]
- 17.Jalbert L, Neill E, Phillips J, Lupo J, et al. Magnetic Resonance Analysis of malignant transformation in recurrent glioma. Neuro Oncology. 2016;18:1169–79. doi: 10.1093/neuonc/now008. E-pub Feb 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu J, Qin L, Cheng S, et al. Residual low ADC and high FA at the resection margin correlate with poor chemoradiation response and overall survival in high-grade glioma patients. European Journal of Radiology. 2016;85(3):657–664. doi: 10.1016/j.ejrad.2015.12.026. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Chen AP, Crane JC, Chang SM, Vigneron DB, Nelson SJ. Three-dimensional J-resolved H-1 magnetic resonance spectroscopic imaging of volunteers and patients with brain tumors at 3T. Magn Reson Med. 2007;58(5):886–92. doi: 10.1002/mrm.21415. [DOI] [PubMed] [Google Scholar]
- 20.Oh J, Henry RG, Pirzkall A, Lu Y, Li X, Catalaa I, Chang S, Dillon WP, Nelson SJ. Survival Analysis in patients with glioblastoma multiforme: predictive value of choline-to-N-acetylaspartate index, apparent diffusion coefficient, and relative cerebral blood volume. J Magn Reson Imaging. 2004;19(5):546–554. doi: 10.1002/jmri.20039. [DOI] [PubMed] [Google Scholar]
- 21.Saraswathy S, Crawford FW, Lamborn KR, Pirzkall A, Chang S, Cha S, Nelson SJ. Evaluation of MR markers that predict survival in patients with newly diagnosed GBM prior to adjuvant therapy. J Neurooncol. 2009;91(1):69–81. doi: 10.1007/s11060-008-9685-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Jin H, Lu Y, Oh J, Chang S, Nelson SJ. Identification of MRi and 1H MRSI parameters that may predict survival for patients with malignant gliomas. NMR Biomed. 2004;17(1):10–20. doi: 10.1002/nbm.858. [DOI] [PubMed] [Google Scholar]
- 23.Hattingen E, Raab P, Franz K, Lanfermann H, Setzer M, Gerlach R, Zanella E, Pilatus U. Prognostic value of choline and creatine in WHO grade II gliomas. Neuroradiology. 2008;50(9):759–767. doi: 10.1007/s00234-008-0409-3. [DOI] [PubMed] [Google Scholar]
- 24.Park I, Chen AP, Zierhut ML, Ozturk-Isik E, Vigneron DB, Nelson SJ. Implementation of 3T lactate-edited 3D 1H MR spectroscopic imaging with flyback echo-planar readout for gliomas patients. Ann Biomed Eng. 2011;39:193–204. doi: 10.1007/s10439-010-0128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKnight TR, Noworolski SM, Vigneron DB, Nelson SJ. An automated technique for the quantitative assessment of 3D-MRSI data from patients with glioma. Journal of Magnetic Resonance Imaging. 2001;13(2):167–177. doi: 10.1002/1522-2586(200102)13:2<167::aid-jmri1026>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 26.Wen P, Macdonald D, Reardon D, Cloughesy T, et al. Updated Response Assessment Criteria for High-Grade Gliomas: Response Assessment in Neuro-Oncology Working Group. J Clin Onc. 2010;28(11):1973–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 27.Khayal IS, McKnight TR, McGue C, Vandenberg S, Lamborn KR, Chang SM, Cha S, Nelson SJ. Apparent diffusion coefficient and fractional anisotropy of newly diagnosed grade II gliomas. NMR Biomed. 2009;22:449–55. doi: 10.1002/nbm.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bian W, Khayal IS, Lupo JM, McGue C, Vandenberg S, Lamborn KR, Chang SM, Cha S, Nelson SJ. Multiparametric characterization of grade 2 glioma subtypes using magnetic resonance spectroscopic, perfusion, and diffusion imaging. Transl Oncol. 2009;2:271–80. doi: 10.1593/tlo.09178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.An Z, Ganji SK, Tiwari V, Pinho MC, Patel T, Barnett S, Pan E, Mickey BE, Maher EA, Choi C. Detection of 2-hydroxyglutarate in brain tumors by triple-refocusing MR spectroscopy at 3T in vivo. Magn Reson Med. 2016 doi: 10.1002/mrm.26347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Park I, Nelson SJ. Imaging tumor metabolism using in vivo magnetic resonance spectroscopy. Cancer J. 2015;21:123–8. doi: 10.1097/PPO.0000000000000097. [DOI] [PMC free article] [PubMed] [Google Scholar]