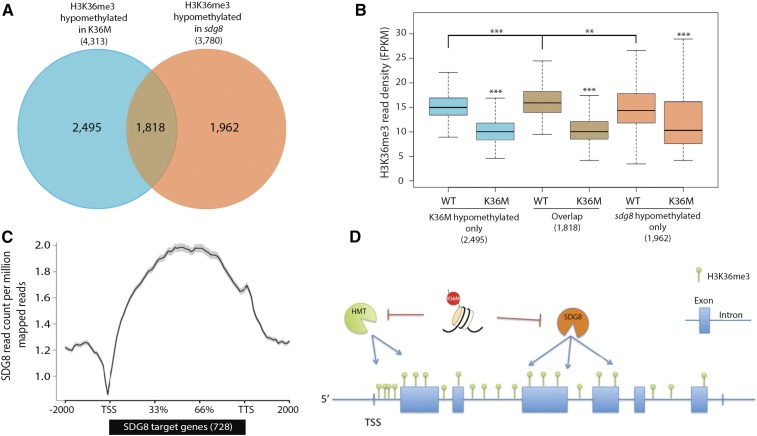
Figure 5.
K36M hypomethylated genes partially overlap with SDG8 targets. A, Overlap of H3K36me3 hypomethylated genes in K36M and sdg8 (hypergeometric P value = 7.58 × 10−315). B, Box plot of H3K36me3 reads over genes only hypomethylated in K36M, overlap, and only hypomethylated in sdg8. C, Metaplot shows that SDG8 primarily binds to the body of its 728 target genes. ChIP-seq of SDG8 is from Li et al. (2015). D, A working model for K36M action. K36M mutant histones likely act via a trans-mechanism to inhibit SDG8 activity and cause the reduction of H3K36me3 in the gene body regions, where SDG8 primarily functions. K36M may also inhibit an unknown histone methyltransferase), which leads to the reduction of H3K36me3 at the TSS. HMT, Methyltransferase; TTS, transcription terminal site. Student’s t test was used to calculate the P values (***P < 0.001, **P < 0.01).