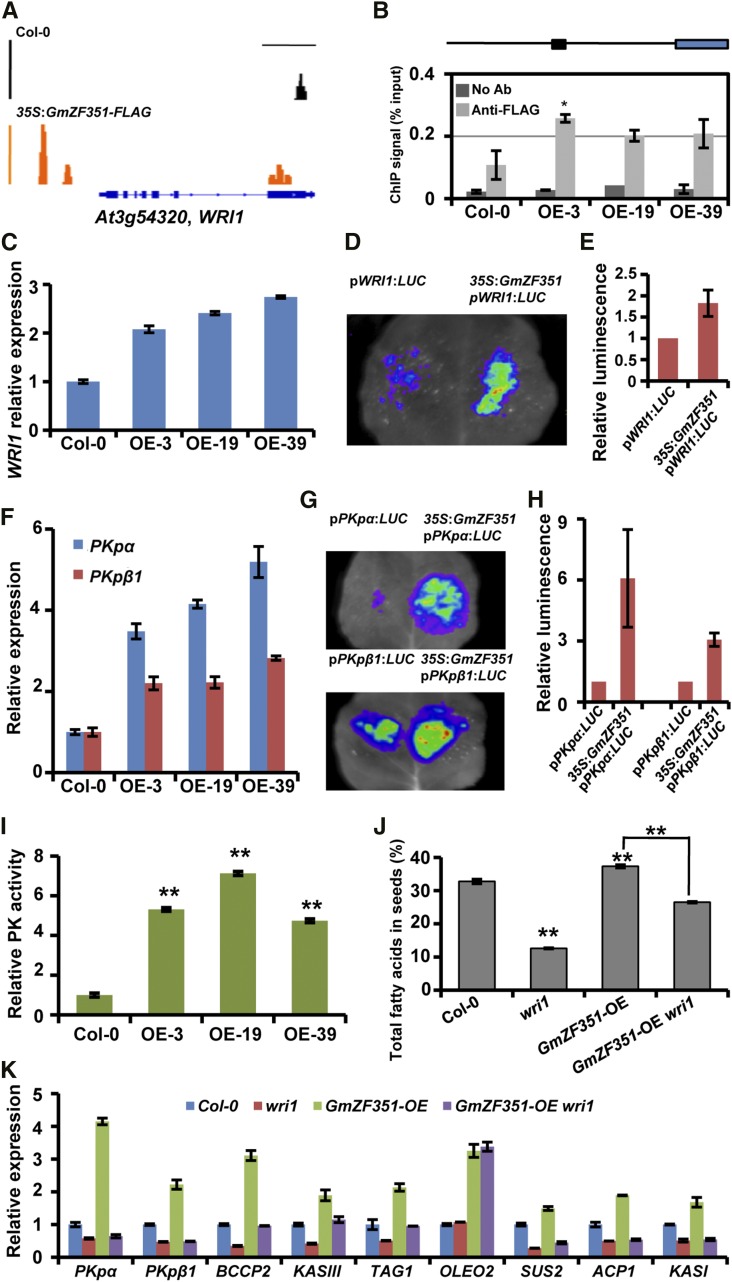
Figure 5.
GmZF351 directly activates WRI1 and WRI1-regulated gene expression. A, ChIP-seq genome browser views of GmZF351 occupancy at the WRI1 locus in Col-0 and 35S:GmZF351-FLAG transgenic plants. B, The WRI1 promoter region is enriched by ChIP-qPCR in GmZF351 transgenic plants. The schematic diagram represents the location of the promoter fragment (black rectangle) used for ChIP-qPCR analysis in the 2-kb promoter region of WRI1 (black line). The blue rectangle indicates the CDS region of WRI1. No Ab samples were immunoprecipitated without antibody and served as a negative control, while isolated chromatin before immunoprecipitation was used as an input control. The ChIP signal (% input) was quantified as a percentage of input DNA by qPCR. Error bars indicate sd (n = 3). The asterisk indicates a significant difference of anti-FLAG ChIP signal in GmZF351-OE plants compared with those in Col-0 (*, P < 0.05). C, RT-qPCR analyses of WRI1 expression. RNA samples were prepared from 5-d-old seedlings. The WRI1 mRNA level relative to At4g12590 in Col-0 was set to 1. Error bars indicate sd (n = 3). D, GmZF351 activates WRI1 promoter activity by transient expression assay in tobacco leaves. A. tumefaciens harboring pWRI1:LUC was mixed with A. tumefaciens harboring 35S:GmZF351-FLAG and infiltrated into tobacco leaves. A. tumefaciens harboring pGWB412 was used as a negative control. The LUC image was taken 2 d after infiltration. E, Quantitative analysis of the luminescence intensity in D. Luminescence intensity was analyzed with IndiGo software. Error bars indicate sd (n = 5). F, RT-qPCR analyses of PKpα and PKpβ1 expression in Col-0 and GmZF351 transgenic Arabidopsis lines. Others are as in C. G, GmZF351 activates PKpα and PKpβ1 promoter activity by transient expression assay in tobacco leaves. A. tumefaciens harboring pPKpα:LUC or pPKpβ1:LUC was mixed with A. tumefaciens harboring 35S:GmZF351-FLAG and infiltrated into tobacco leaves. Others are as in D. H, Quantitative analysis of the luminescence intensity in G. Others are as in E. I, Pyruvate kinase (PK) activity of Col-0 and GmZF351 transgenic Arabidopsis plants. Pyruvate kinase activity in Col-0 was set to 1. Error bars indicate sd (n = 4). Asterisks indicate significant differences from Col-0 (**, P < 0.01). J, Total FA contents in seeds of Col-0, wri1, GmZF351-OE, and GmZF351-OE wri1 plants. Error bars indicate sd (n = 4). The values from GmZF351-OE wri1 were only compared with GmZF351-OE. Asterisks indicate significant differences compared with Col-0 or between the compared pairs (**, P < 0.01). K, Expression levels of PKpα, PKpβ1, BCCP2, KASIII, TAG1, OLEO2, SUS2, ACP1, and KASI in 5-d-old seedlings of Col-0, wri1, GmZF351-OE (OE-19), and GmZF351-OE wri1 plants. The mRNA levels (relative to At4g12590) of each gene in Col-0 were set to 1. Error bars indicate sd (n = 3).