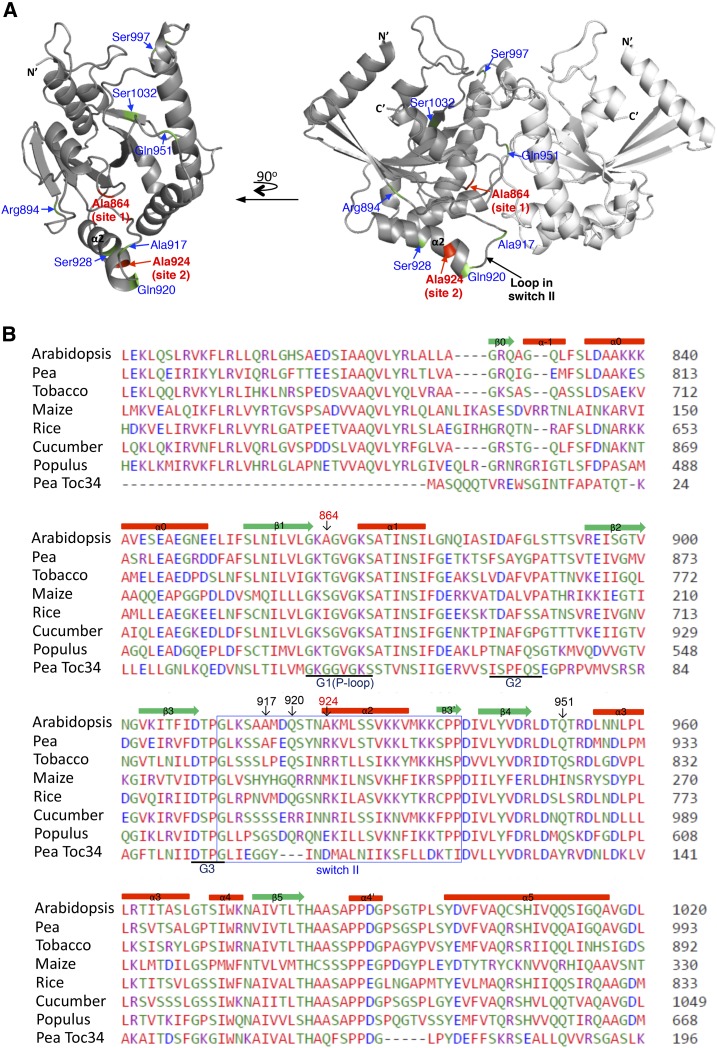
Figure 4.
AtToc159G structure model and sequence alignment. A, The two deduced sites cleaved by FeBABE-conjugated preproteins are colored in red. Additional residues mutated to Cys are indicated and colored in green. The N- and C-terminal ends, α-helix 2, and the central loop in switch II also are indicated. At left, one of the monomers is rotated 90° horizontally to show the dimer interface. B, Sequence alignment of the G domain region of Toc159 homologs from representative higher plant species and the pea Toc34 G domain. Secondary structure elements are marked above the sequence, and G1 to G3 motifs are marked below the sequence, all of which are labeled according to the pea Toc34 crystal structure (Protein Data Bank no. 1h65). The predicted switch II is boxed. Deduced residues cleaved by FeBABE and residues that generated significant cross-linking to preproteins when mutated to Cys are indicated with red and black numbers, respectively. Accession numbers for the sequences shown are as follows: At4g02510 (Arabidopsis Toc159), AAF75761 (pea Toc159), XP_009769991 (tobacco [Nicotiana tabacum] Toc159), NP_001147969 (maize [Zea mays] Toc159), XP_015639221 (rice [Oryza sativa] Toc159), XP_004152365 (cucumber [Cucumis sativus] Toc159), XP_002312975 (Populus spp. Toc159), and Q41009 (pea Toc34).