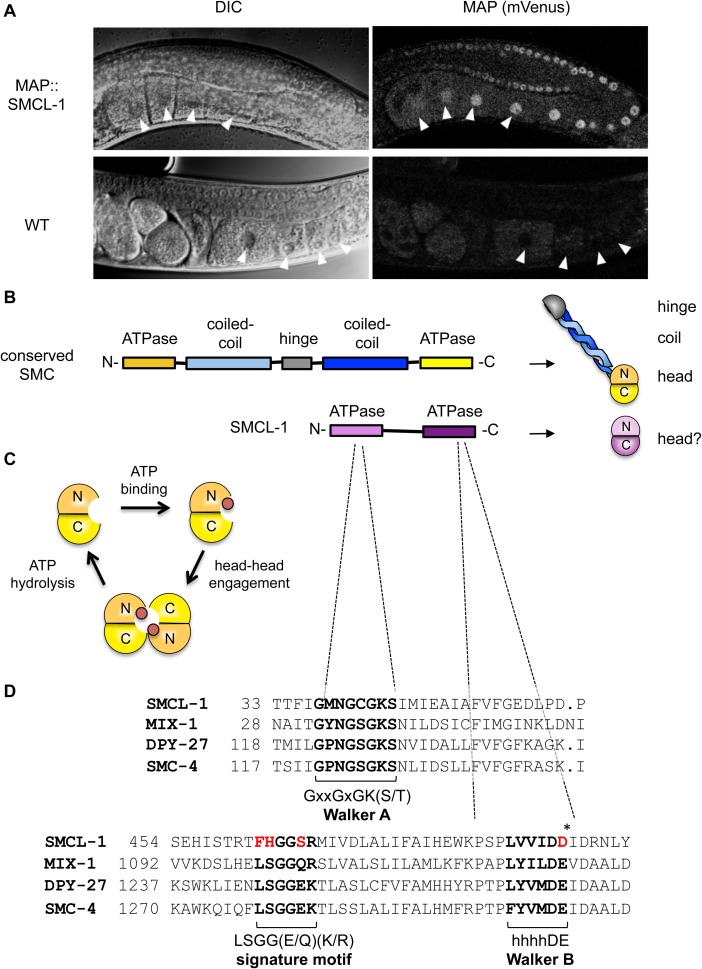
Fig 3. SMCL-1 expression and protein features.
(A) Adult hermaphrodites from wild-type (WT) and a strain carrying the map::smcl-1 transgene driven by endogenous smcl-1 5’ and 3’ elements. A section of the germline is shown, imaged by DIC to show structures and fluorescent microscopy to detect mVenus expression from the MAP tag. Arrowheads denote the first four oocytes. (B) A typical SMC protein folds back on itself at a hinge domain, bringing coil regions together and creating a “head domain” (yellow) from ATPase domains in the N- and C-termini. SMCL-1 lacks predicted coil and hinge domains, but has N- and C-terminal ATPase domains that may be capable of forming a head domain (purple). (C) SMC head domain and the ATPase cycle, showing binding of ATP (red circle), ATP-dependent engagement of heads from two SMC proteins, and disengagement upon ATP hydrolysis. (D) SMCL-1 amino acid sequence aligned to C. elegans condensin SMC proteins. Shown are regions surrounding three conserved motifs found in SMCs and related ATPases: the Walker A motif, ABC transporter signature motif, and Walker B motifs, and their consensus sequences. SMCL-1 shares a conserved Walker A motif, but differs from consensus signature motif and Walker B motif at residues shown in red. Asterisk denotes catalytic amino acid required for ATP hydrolysis. x = any amino acid and h = hydrophobic amino acid.