Figure 3. Physiological Data for Experiment 3.
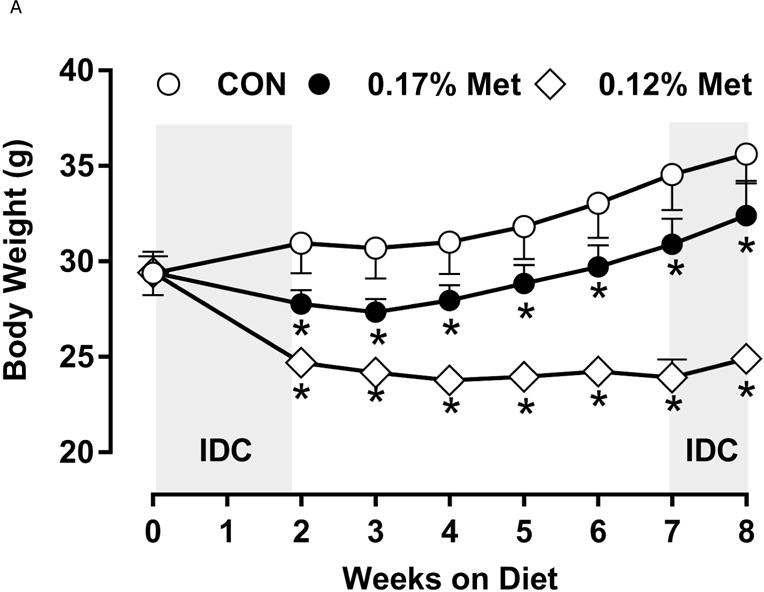
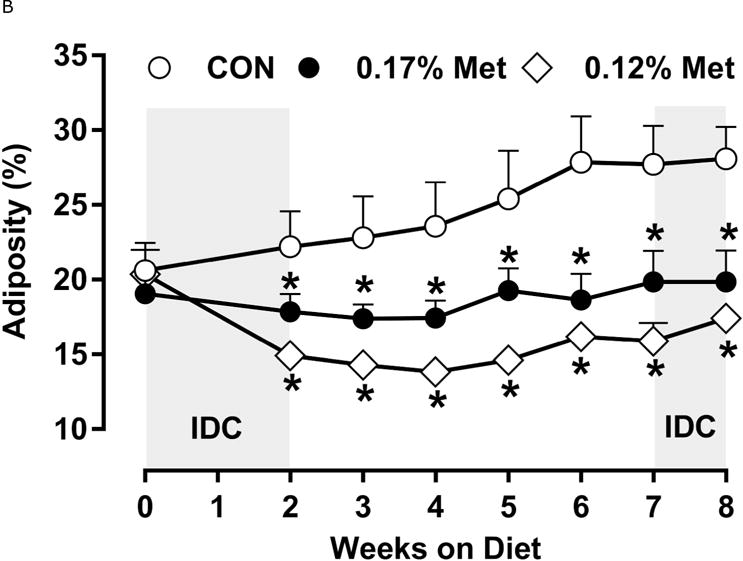
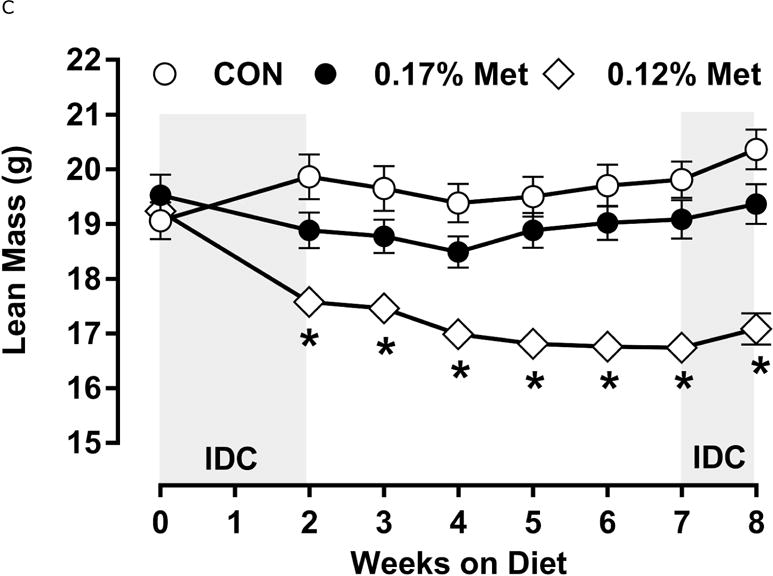
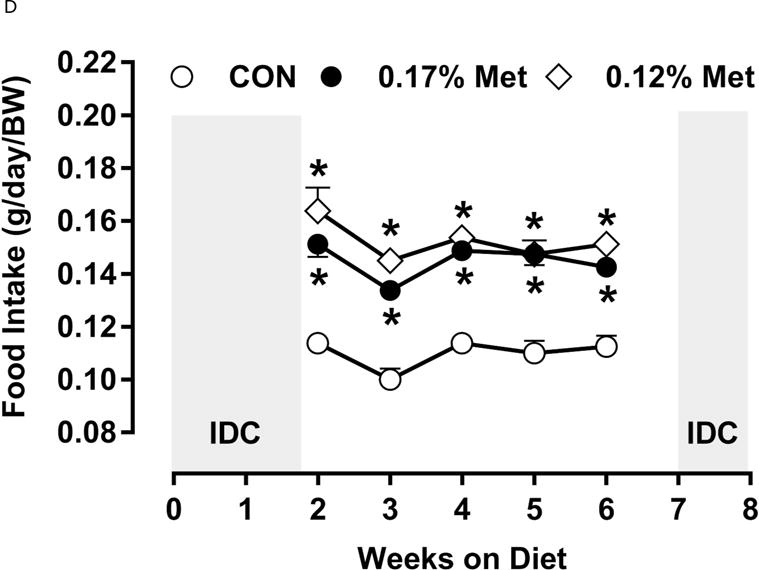
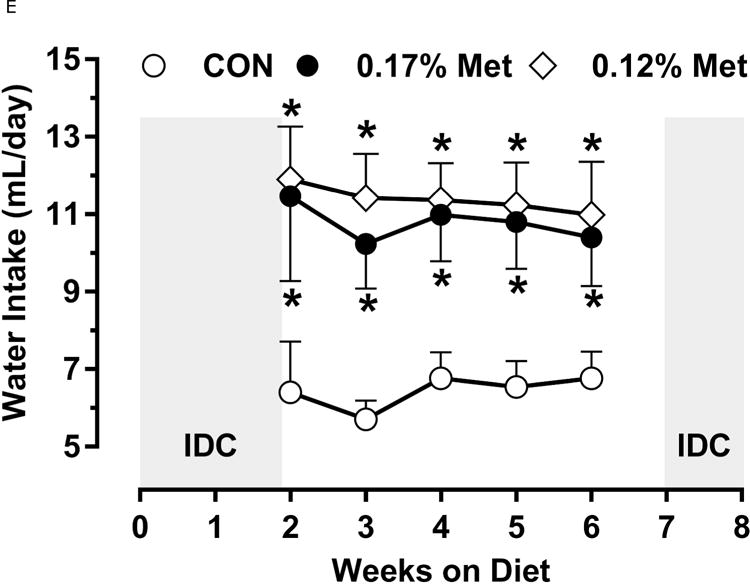
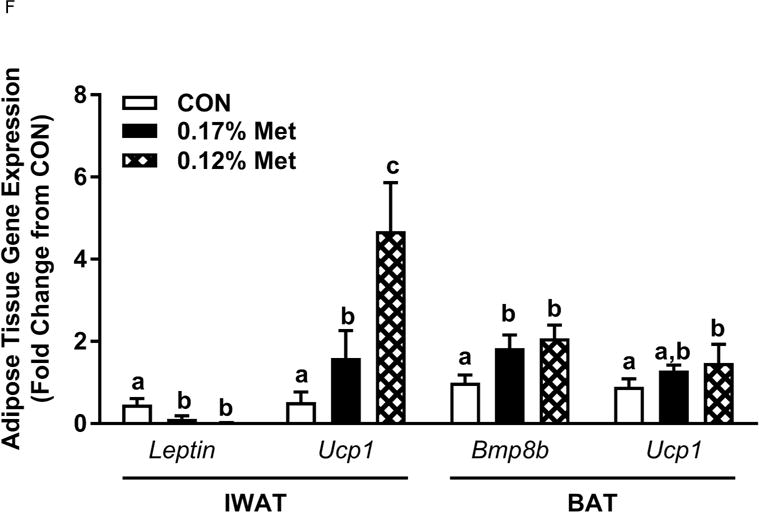
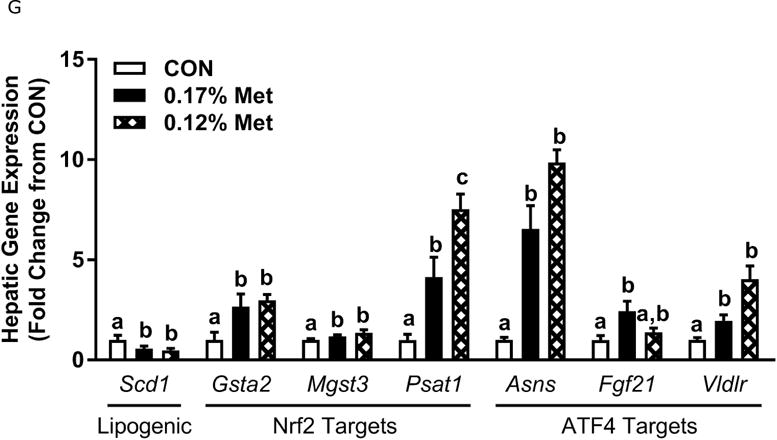
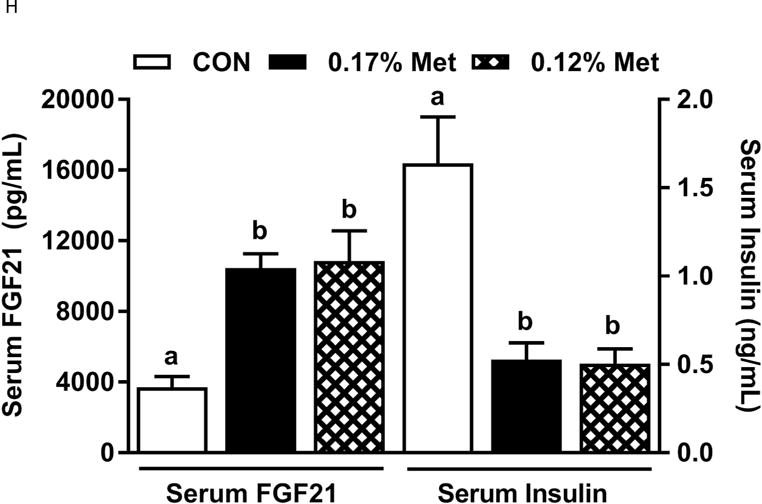
Body weight (A), adiposity (B), lean mass (C), food intake (D), and water intake (E) were measured at weekly intervals over the course of the study. Expression of selected genes in adipose tissue (F) and liver (G) were measured via qPCR and expressed as the fold change of the MR group over the CON group. Serum metabolites FGF21 and insulin (H) were measured via ELISA. CON – control; MR – Met restricted diets; IDC – indirect calorimetry; IWAT – inguinal white adipose tissue; BAT – brown adipose tissue; Scd1 – stearoyl CoA desaturase 1; Nrf2 – Nuclear factor erythroid 2-related factor 2; Gsta2 – Glutathione S-Transferase α2; Mgst3 – Microsomal Glutathione S-Transferase 3; Psat1 – Phosphoserine Aminotransferase 1; ATF4 – Activating Transcription Factor 4Asns – asparagine synthase; Vldlr – very low density lipoprotein receptor; Fgf21 – fibroblast growth factor 21; Ucp1 – uncoupling protein 1; Bmp8b – bone morphogenetic protein 8b. All values are expressed as mean ± SEM. Means annotated with an asterisk differ from CON at p < 0.05. In panels F and G, means not sharing a common letter differ at p < 0.05.
