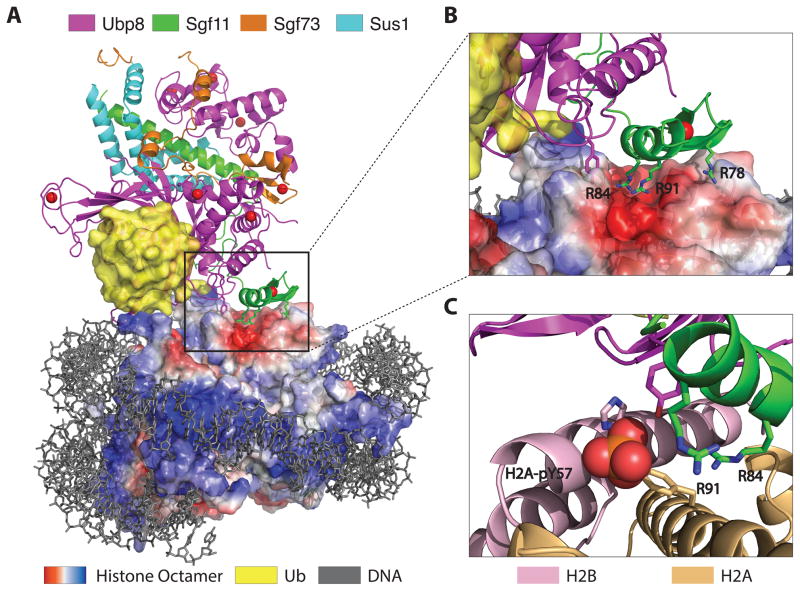
Figure 1.
Docking of the SAGA DUB module on H2B-ubiquitinated nucleosomes. (A) Structure of the yeast DUB module (Ubp8, Sgf11, Sgf73, Sus1) bound to a nucleosome core particle containing monoubiquitinated H2B. Surface representation of the histone octamer shows negative (red) and positive (blue) electrostatic potential. (B) Docking of the arginine-rich Sgf11 zinc finger (green) on the acidic cleft (red) between H2A and H2B. (C) Modeling showing position of phosphorylated H2A-Y57 at the interface between the DUB module and histones H2A and H2B.