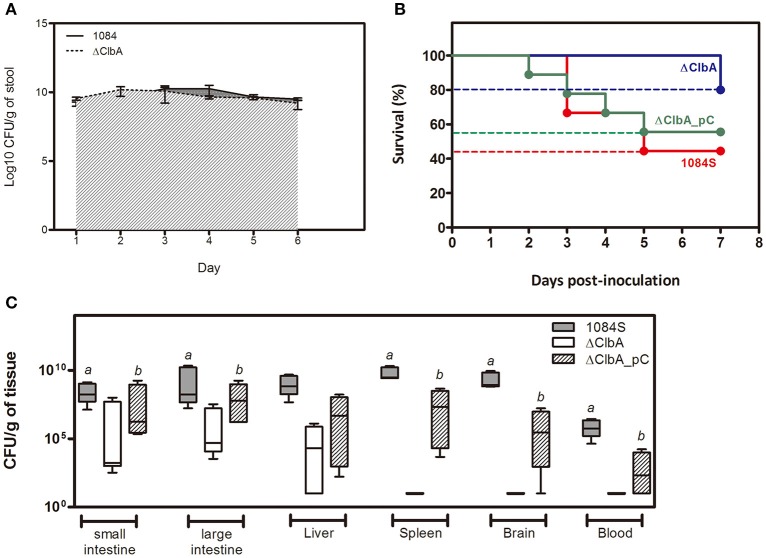
Figure 1.
Involvement of colibactin in intestinal colonization and the capacity of K. pneumoniae 1084, a pks+ K1 CC23 hypervirulent strain, to establish systemic infections. Groups of 8-wk old BALB/c male mice were orogastrically inoculated with 1 × 109 CFU of K. pneumoniae 1084S, ΔClbA, or ΔClbA_pC. (A) Fecal shedding of 1084S (solid line) and ΔClbA (slash line) presented as a geometric mean of CFU per gram of stool, determined daily by a plate count method. (B) Survival rates of 1084S- (n = 9; red line), ΔClbA- (n = 6; blue line), or ΔClbA_pC-inoculated (n = 9; green line) mice determined by Kaplan-Meier analysis using Prism5 (GraphPad) are shown. (C) At the 7th day post-inoculation, the infected mice which survived the experimental period were sacrificed. The mucosa of the small and large intestines, liver, spleen, brain, and blood were retrieved and homogenized for the determination of bacterial loads for the 1084S- (dark gray), ΔClbA- (white), and ΔClbA_pC- (slash) group. CFU per gram of tissues is presented in a box and whisker plot. The edges of each box are the 25th and 75th percentiles, and the middle is the median. Lower case letter “a” and “b” represents statistical significance of CFUg−1, p < 0.05 (one-tailed) determined by Mann-Whitney test, between 1084S/ΔClbA and ΔClbA/ΔClbA_pC, respectively.