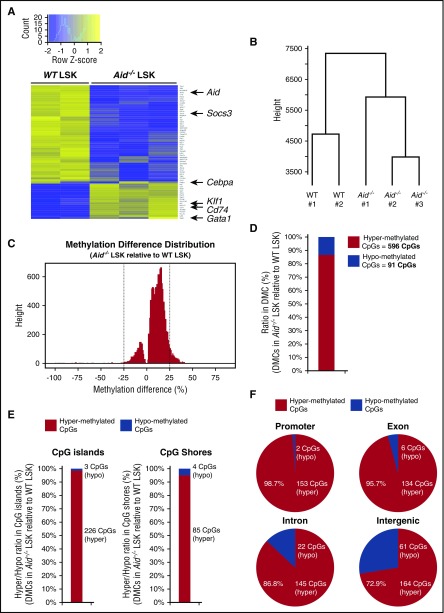
Figure 6.
Deletion of Aid leads to transcriptional alteration of some key regulators of erythropoiesis and enhanced DNA methylation in gene regulatory elements. (A) Differential gene expression analysis from RNA-seq data of WT and Aid−/− BM LSK cells (WT, n = 2; Aid−/−, n = 3). (B) Dendrogram of the hierarchical clustering based on RNA-seq profiles from WT and Aid−/− LSK cells (WT, n = 2; Aid−/−, n = 3). (C) Methylation difference distribution of CpGs (q < 0.01 only) in Aid−/− LSK cells relative to WT LSK cells based on eRRBS methylation profiles. Pattern clearly shows shift toward more hypermethylated phenotype. DMCs were called as the CpGs with q < 0.01 and >25% change in either direction (as shown by the vertical lines). (D) The ratio and the exact number of hypermethylated and hypomethylated CpGs within total DMCs in Aid−/− LSK cells relative to WT LSK cells. (E) The ratio and the exact number of hypermethylated and hypomethylated CpGs within DMCs localized in CpG islands or CpG shores in Aid−/− LSK cells relative to WT LSK cells. (F) The ratio and the exact number of hypermethylated and hypomethylated CpGs within DMCs localized in coding regions in Aid−/− LSK cells relative to WT LSK cells.