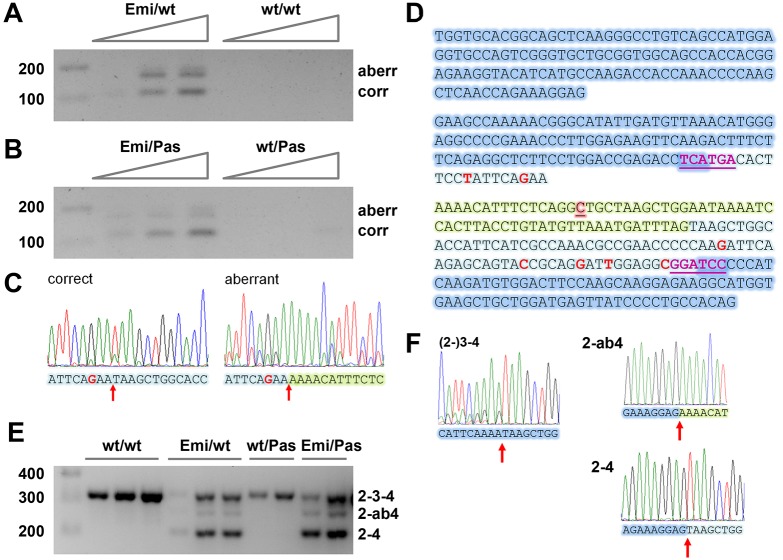
Fig. 2.
Alternative splicing of transcripts from the humanized Fech allele in transgenic mice. (A,B) RT-PCR analysis (40 cycles) with primers specific for the humanized allele of increasing amounts of total liver RNA from (A) Emi/wt and wt/wt C57BL/6J mice and (B) Emi/Pas and wt/Pas F1 hybrid mice. Both correctly and aberrantly spliced transcripts are detected. A band migrating slightly above the aberrant one is due to heterodimer formation. (C) Sanger sequence traces of PCR-amplified cDNA corresponding to correct and aberrant transcripts. The splice junctions are marked by a red arrow. (D) Sequence of exons 2 to 4 of the humanized allele. The shading indicates mouse exon parts (dark blue), human exon parts (light blue) and the extra 63 nucleotides added as a result of aberrant splicing (light green). Within the exons, single nucleotide differences between human and mouse are marked in red (all silent changes not affecting the protein sequence). The c.315-48C polymorphism is highlighted in red and underlined. Magenta, underlined letters: BspHI and BamHI sites in exons 3 and 4. (E) RT-PCR analysis (35 cycles) with primers in the common parts of exons 2 and 4 of total spleen RNA from multiple mice of the indicated genotypes. (F) Sanger sequence traces demonstrating that the bands correspond to full-length Fech mRNA [(2-)3-4], as well as exon 3-skipped mRNA spliced to the aberrant and correct 3′ ss (2-ab4 and 2-4, respectively). For primer sequences, see Table S1.