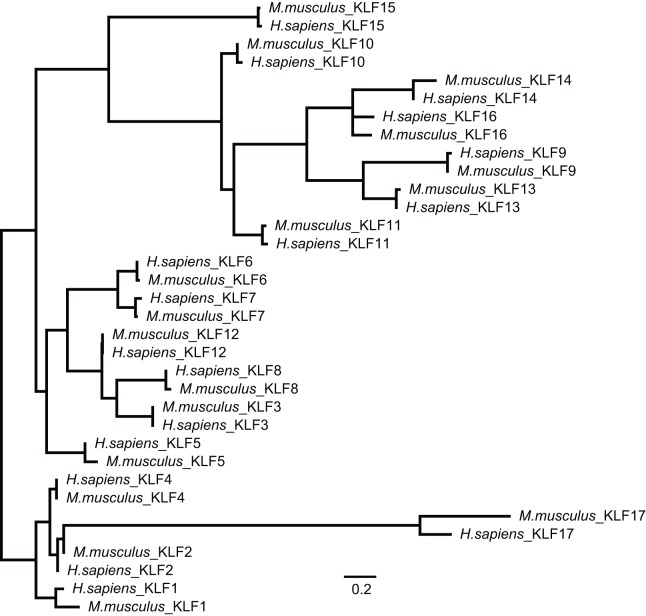
Fig. 2.
Phylogenetic relationship between human and mouse KLF family members. An amino acid alignment was produced from full-length mouse and human KLF family members using MAFFT (Katoh et al., 2002) employing the L-INS-i algorithm and BLOSUM45 scoring matrix. As KLF family members share little homology outside of the C-terminal ZNF region, the alignment was trimmed 22 residues N-terminal of the first ZNF domain and one residue C-terminal of the third ZNF domain. Phylogenetic analysis of the ZNF region was performed via RAxML 7.2.8 (Stamatakis, 2006) using the Gamma LG model, and node support in the ML tree was sampled via 100 bootstrap replicates. Both alignment and phylogenetic analyses were performed in Geneious version 8.1.8 (Kearse et al., 2012). Scale bar: 0.2 amino acid changes per site.