SUMMARY
Genetic S6K1 inactivation can induce apoptosis in PTEN-deficient cells. We analyzed the therapeutic potential of S6K1 inhibitors in PTEN-deficient T cell leukemia and glioblastoma. Results revealed that the S6K1 inhibitor LY-2779964 was relatively ineffective as a single agent, while S6K1-targeting AD80 induced cytotoxicity selectively in PTEN-deficient cells. In vivo, AD80 rescued 50% of mice transplanted with PTEN-deficient leukemia cells. Cells surviving LY-2779964 treatment exhibited inhibitor-induced S6K1 phosphorylation due to increased mTOR-S6K1 co-association, which primed rapid recovery of S6K1 signaling. In contrast, AD80 avoided S6K1 phosphorylation and mTOR co-association, resulting in durable suppression of S6K1-induced signaling and protein synthesis. Kinome analysis revealed that AD80 coordinately inhibits S6K1 together with the TAM family tyrosine kinase AXL. TAM suppression by BMS-777607 or genetic knockdown potentiated cytotoxic responses to LY-2779964 in PTEN-deficient glioblastoma cells. These results reveal that combination targeting of S6K1 and TAMs is a potential strategy for treatment of PTEN-deficient malignancy.
Keywords: S6K, S6K1, rapamycin, apoptosis, Leukemia, kinase inhibitor, DG2, LY-2779964, PF4708671, AD80, cancer, chemotherapy, Pten, BMS-777607, AXL, TAM, glioblastoma
INTRODUCTION
Inactivation of the phosphatidylinositol 3′-phosphatase PTEN as a result of genomic mutation, epigenetic silencing, and/or non-coding RNA regulation is a frequent event in glioblastoma and T acute lymphoblastic leukemia (Cerami et al., 2012; Gao et al., 2013; Gutierrez et al., 2009; Song et al., 2012, Cancer Genome Atlas Research, 2008 #1422). Through Akt-mTORC1 signaling, PTEN loss triggers the hyperactivation of ribosomal protein S6 kinase 1 (S6K1). Activated S6K1 promotes protein synthesis through a variety of mechanisms, including increased translation initiation through the phosphorylation of eIF4B and PDCD4 (Dorrello et al., 2006; Raught et al., 2004; Shahbazian et al., 2010). S6K1 also regulates translation in a non-catalytic manner through its phosphorylation-dependent association with the eIF3 complex (Holz et al., 2005). Thus S6K1 is a hub for translation control downstream of mTORC1.
We previously showed that genetic inactivation of S6K1 in PTEN-deficient hematopoietic cells reduced glucose-dependent cell survival and significantly delayed the incidence of leukemia in vivo (Tandon et al., 2011). In a parallel study, the absence of S6K1 reduced the incidence of adrenal tumors in PTEN+/− mice (Nardella et al., 2011). These results indicated that development of S6K1 targeted therapeutics would be beneficial for treatment of PTEN-deficient malignancy.
Recently a number of S6K1 inhibitor compounds have become available. The polykinase inhibitor DG2 has been used to inhibit S6K1 in several studies of translation control (Hsieh et al., 2010; Okuzumi et al., 2009; Wang et al., 2011). PF-4708671 has been used to investigate S6K1 function in glioblastoma survival signaling (Gruber Filbin et al., 2013) and the regulation of pyrimidine biosynthesis (Ben-Sahra et al., 2013; Robitaille et al., 2013). The compound LY-2779964 (LY-2584702 tosylate) was recently described in a single agent Phase I trial in patients with advanced cancers (Tolcher et al., 2014). In parallel, the polykinase inhibitors AD57 and AD80 were shown to inhibit S6K1 and suppress oncogenic function downstream of a transforming mutant of the receptor tyrosine kinase Ret (Dar et al., 2012). Here we analyze the efficacy of these inhibitors in PTEN-deficient malignant cells, revealing S6K1 as a key component of a multikinase targeting strategy that is selectively cytotoxic in PTEN-deficiency.
RESULTS
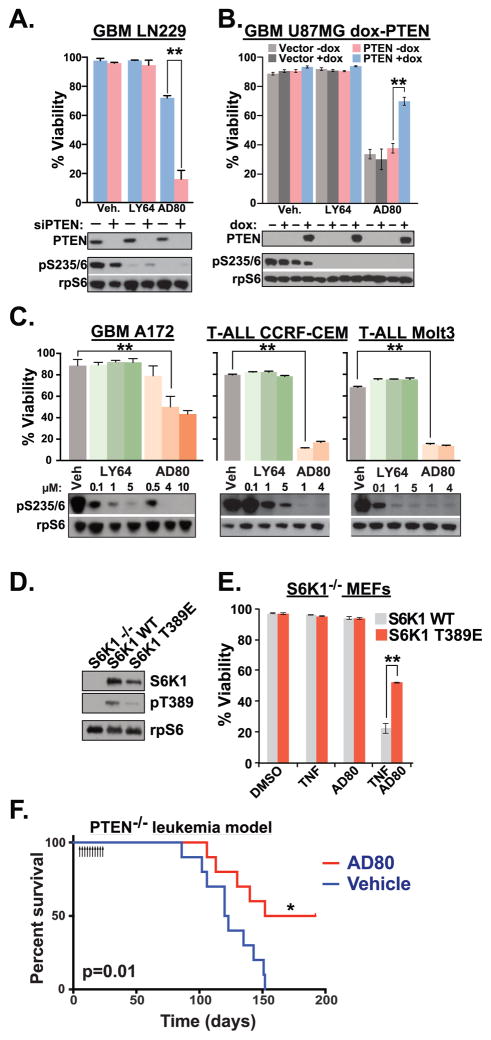
We investigated the cytotoxic effects of recently described S6K1 inhibitors AD80 and LY-2779964. LY-2779964 is the tosylate salt of LY-2584702, which has been previously described in a Phase I trial for patients with advanced solid tumors (Tolcher et al., 2014). In LN229 and GAMG glioblastoma cells treated with either non-targeting or PTEN-targeting siRNA (siNT and siPTEN, respectively), both AD80 and LY-2779964 (LY64) were effective in reducing the S6K1-dependent phosphorylation of the ribosomal protein S6 (rpS6) at 3 hours (Figures 1a, S1a). However, only AD80, and not LY-2779964, reduced the viability of PTEN-knockdown cells. In U87 PTEN-deficient glioblastoma cells, inducible PTEN reexpression rendered cells relatively resistant to the effects of AD80 (Figure 1b). Again, LY-2779964 was ineffective in inducing cytotoxic responses. In PTEN-deficient glioblastoma (A172) and T cell leukemia cells (CCRF-CEM and MOLT3), AD80 treatment induced cytotoxic responses while LY-2779964 had little effect (Figure 1c). To genetically analyze determinants of AD80 efficacy, we used gene-targeted murine embryonic fibroblast (MEF) cells. MEFs treated with TNFα in combination with inhibitors of protein synthesis such as cycloheximide (Figure S1b) or AD80 (see below) undergo apoptosis (Lee et al., 2000). Activation of mTORC1-S6K1 via inactivation of PTEN, TSC1, or TSC2 sensitized cells to programmed cell death upon treatment with TNFα in combination with AD80 (Figure S1c,d). In contrast, S6K1−/− MEFs which reexpressed activated S6K1 T389E were resistant to TNFα + AD80, compared to cells reexpressing S6K1 WT (Figure 1d,e). Thus, PTEN-selective cytotoxic targeting of S6K1 is a property of the S6K1 inhibitor AD80 and not LY-2779964.
FIGURE 1.
Pharmacologic targeting of S6K1 in PTEN-deficient cells. (a) PTEN-expressing LN229 glioblastoma cells were transfected with Non-Targeting siRNA (siNT) or siPTEN duplexes, then cultured in 5 μM LY-2779964 (LY64), 10 μM AD80, or vehicle control (Veh). Mean cell viability was determined after 72 hours of culture in indicated conditions (top). Immunoblot of cell lysates from parallel wells after 3 hours demonstrated knockdown of PTEN and inhibition of ribosomal protein S6 (rpS6) phosphorylation (bottom) n=4. (b) PTEN-deficient U87 glioblastoma cells were transduced with vector or doxycycline-regulated PTEN expression constructs. After 48 hours of doxycycline, cells were treated with vehicle, 5 μM LY-2779964, or 10 μM AD80. n=3 viability; n=2 immunoblots. (c) Viability and immunoblot analysis of PTEN-deficient and T-ALL cells treated as in (a). n=4 viability analyses; n=2 immunoblot analyses. (d) Expression of S6K1-WT or S6K1 T389E in S6K1−/− MEFs after viral transduction and selection by sorting. (e) S6K1−/− MEFs reconstituted with WT S6K1 or S6K1 T389E were treated with 30ng/ml TNFα, 10 μM AD80 or combined agents as indicated. S6K1 T389E mediated resistance to TNFα + AD80. n=4. See also Figure S1. (f) CD45.2+ PTEN-deficient bone marrow cells were transplanted into CD45.1+ congenic recipient mice. Groups of 10 mice were injected i.p. with 20 mg/kg AD80 or vehicle control for 10 consecutive days starting at day 5 post transplant, then monitored for signs of leukemia. * p=0.01 Log-Rank test. See also Figure S2. **p<0.01 by two-tailed t-test (a–e).
We investigated the efficacy of AD80 for reducing leukemia in mice that were transplanted with PTEN-deficient bone marrow cells. First we established that AD80 i.p. injection at 20 mg/kg reduced phosphorylated rpS6 in mouse spleen and bone marrow (Figure S2a,b). Next, CD45.2+ donor bone marrow cells were harvested from leukemic Mx1-Cre+; PTENfl/fl mice that had been treated with pIpC to induce PTEN deletion (Yilmaz et al., 2006; Zhang et al., 2006). CD45.1+ recipient mice transplanted with PTEN-deficient leukemic cells were divided into groups for injection of AD80 or vehicle control daily for 10 days. AD80 treatment was tolerated as assessed by tracking of body weight during treatment (Figure S2c). All mice from the vehicle-control group developed leukemia, but only 50% of AD80-treated mice developed leukemia (Figure 1f). AD80-treated mice that did develop leukemia did so with delayed incidence. Thus in vitro cytotoxic effects of AD80 are consistent with in vivo efficacy for PTEN-deficient cancers.
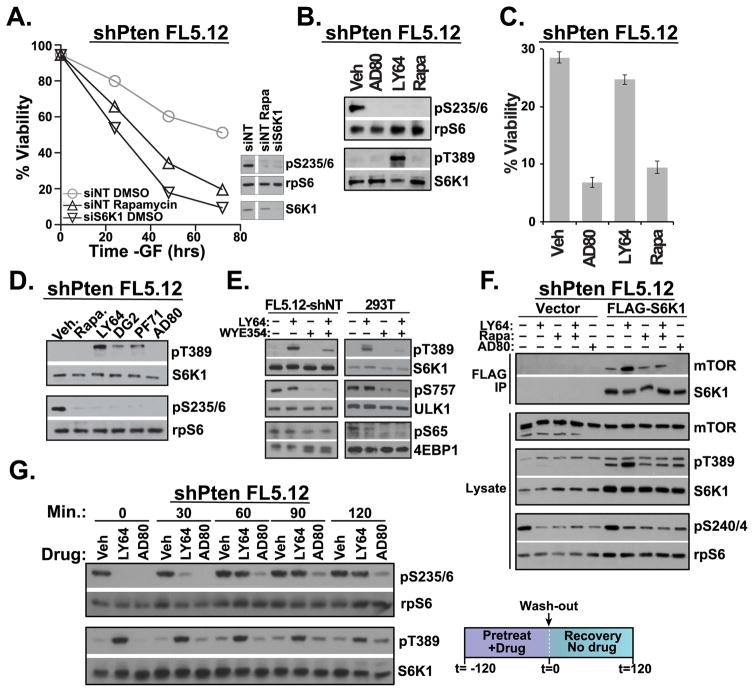
To investigate the mechanistic differences between AD80 and LY-2779964 in targeting S6K1 in PTEN-deficient cells, we determined inhibitor effects in growth factor-dependent FL5.12 hematopoietic progenitor cells. As shown previously (Tandon et al., 2011), PTEN-knockdown is sufficient to mediate growth factor-independent survival that requires active mTORC1-S6K1 signaling (Figures 2a, S3a). Similar to S6K1 knockdown and rapamycin treatment, AD80 and LY-2779964 reduced the phosphorylation of rpS6 (Figure 2b). However, LY-2779964 was only modestly cytotoxic, while AD80 and rapamycin restored apoptosis in PTEN-deficient cells (Figure 2c). We hypothesized that differential biochemical effects contribute to functional differences in S6K1 inhibitors. Indeed, LY-2779964 induced a dramatic increase in the phosphorylation of S6K1 at the hydrophobic motif T389 even while the phosphorylation of downstream rpS6 was inhibited (Figure 2b). In contrast, phosphorylation of S6K1 at T389 was not increased in AD80 treated cells.
FIGURE 2.
S6K1 inhibitor pathway dynamics. (a) IL-3-dependent FL5.12 cells that had been stably transduced with shPTEN were cultured in full medium lacking only IL-3. Transfection of siS6K1 siRNA, or treatment with 20 nM rapamycin induced apoptosis in PTEN-deficient cells. n=3. (b) PTEN-deficient FL5.12 cells cultured –IL-3 for 1 hour in the presence of 4 μM AD80, 1 μM LY-2779964 (LY64) or 20 nM rapamycin (Rapa) revealed reduced rpS6 phosphorylation yet substantial differences in S6K1 T389 phosphorylation. n=2. (c) Viability of cells treated as in (b) for 72 hours. (d) 1 μM LY-2779964, 10 μM DG2, and 10 μM PF-4708671 share an ability to increase pT389 when added to PTEN-deficient FL5.12 cell cultures for 2 hours. 4 μM AD80 avoids the induction of pT389. n=2. (e) shNT FL5.12 cells or serum-starved 293T cells were incubated with 1 μM LY-2779964 or the mTOR ATP-competitive inhibitor WYE354 (1 μM) for 30 minutes. n=4, FL5.12-shNT; n=2, 293T. (f) PTEN-deficient FL5.12 cells transduced with either vector or FLAG-S6K1 were cultured in the indicated inhibitors for 3 hours, prior to lysis and FLAG-IP. n=3. (g) PTEN-deficient FL5.12 cells were preincubated with vehicle control or 1 μM LY-2779964 or 10 μM AD80 for 2 hours. Cells were then washed and phosphorylation kinetics determined by immunoblot. n=3. See also Figure S3.
Increased phosphorylation of S6K1 has been described in cells treated with the S6K inhibitor PF-4708671 (Pearce et al., 2010). We therefore compared phosphorylation of S6K1 in cells treated with a panel of S6K1 inhibitors: PF-4708671, LY-2779964, AD80, and DG2 (described in (Okuzumi et al., 2009)). LY-2779964, DG2, and PF-4708671 reduced substrate rpS6 phosphorylation while inducing substantial phosphorylation of S6K1 at T389 (Figure 2d). In contrast, AD80 suppressed rpS6 phosphorylation and avoided the induction of S6K1 T389 phosphorylation. LY-2779964 mainly affected S6K1 T389, as there were only modest effects on the phosphorylation of S6K1 turn motif S371 and activation loop T229 sites (Figure S3b).
Increased S6K1 T389 phosphorylation upon treatment with LY-2779964, could be related to a general increase in mTORC1 activity. However, increased S6K1 T389 phosphorylation was not accompanied by a general increase in the phosphorylation of the parallel mTORC1 substrates 4EBP1 and ULK1 (Figure 2e). The mTOR-dependence of these phosphorylation sites is confirmed by treatment with the mTOR catalytic site inhibitor WYE-354. Combination of WYE-354 with LY-2779964 substantially reduced S6K1 phosphorylation at pT389 (Figure 2e, see also Figure S3b). Similarly, knockdown of the mTORC1 specific subunit raptor reduced phosphorylation of S6K1 at T389 (Figure S3c). Further analysis by co-immunoprecipitation indicated that LY-2779964 induced the association of S6K1 with mTOR (Figure 2f). Altogether the results favor a model in which LY-2779946 induces the association of S6K1 with mTOR, mediating mTORC1-dependent T389 phosphorylation. Additional mechanisms including effects on inhibitor-induced dimerization may also contribute to increased S6K1 pT389.
To determine the effects of inhibitor-induced kinase phosphorylation in signal transduction, we examined the kinetics of substrate and kinase phosphorylation after drug washout. After preincubation, S6K1 T389 phosphorylation was substantially increased by LY-2779964 treatment while rpS6 phosphorylation was reduced (Figure 2g). rpS6 phosphorylation rebounded within 60 minutes after removal of LY-2779964, while there was an extended delay rpS6 phosphorylation in AD80-treated cells. Together these results reveal substantial differences in kinase phosphorylation and signaling kinetics between LY-2779964 and AD80, which correspond to differential cytotoxic efficacy.
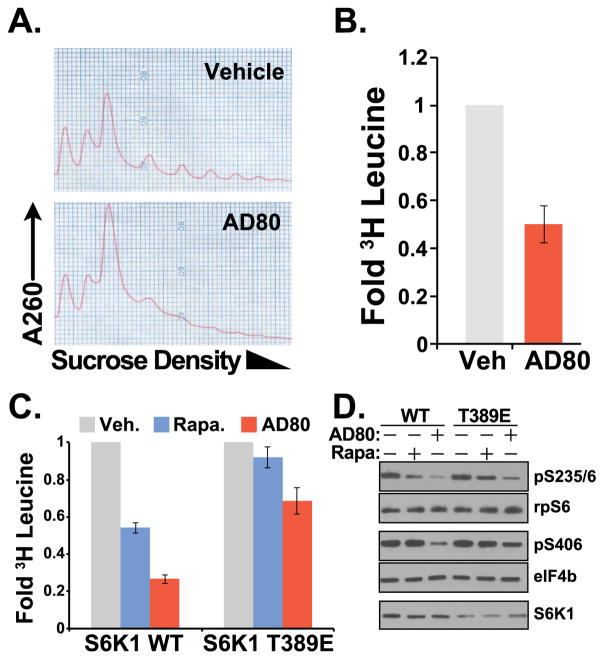
The anti-neoplastic activity of S6K1 knockout/knockdown has been linked to the regulation of new protein synthesis in APC-deficient colorectal cancer cells (Faller et al., 2015). In PTEN-deficient cells, AD80 treatment dramatically reduced the fraction of polysome-associated mRNA (Figure 3a), suppressing the incorporation of 3H-leucine into newly synthesized proteins (Figure 3b). To determine the effects of AD80 on S6K1 control of protein synthesis, we measured the phosphorylation of the S6K1 substrates rpS6 and eIF4B in S6K1−/− MEFs that reexpressed WT S6K1 or the S6K1 T389E activated mutant. S6K1 T389E substantially protected the phosphorylation of rpS6 in cells treated with rapamycin, which corresponded with an increase in new protein synthesis (Figure 3c,d). AD80 mediated a striking reduction in protein synthesis in cells expressing S6K1 WT, which corresponded in reduced phosphorylation of rpS6 and eIF4b. Protein synthesis and substrate phosphorylation was substantially protected from AD80 by the expression of S6K1 T389E (Figure 3c,d).
FIGURE 3.
Targeting translation through S6K1. (a) rRNA polysome assembly in PTEN-deficient FL5.12 cells cultured –growth factor for 3 hours ± 4 μM AD80, n=4. (b) 3H-leucine incorporation into the TCA-precipitated protein fraction from PTEN-deficient FL5.12 cells treated ±4 μM AD80. Measurements are mean±SD cpm from triplicate wells cultured in parallel and standardized to Vehicle control. n=5. (c) S6K1−/− MEFs reconstituted with WT-S6K1 or S6K1 T389E were incubated with vehicle control, 20 nM rapamycin or 4 μM AD80. Measurements are as in (b), n=4. (d) Immunoblot analysis of cells cultured as in (c) for 2 hours reveals that S6K1 T389E sustained phosphorylation of the S6K1 substrates rpS6 and eIF4b, indicating a mechanism for AD80 regulation of S6K1-dependent translation control; n=3.
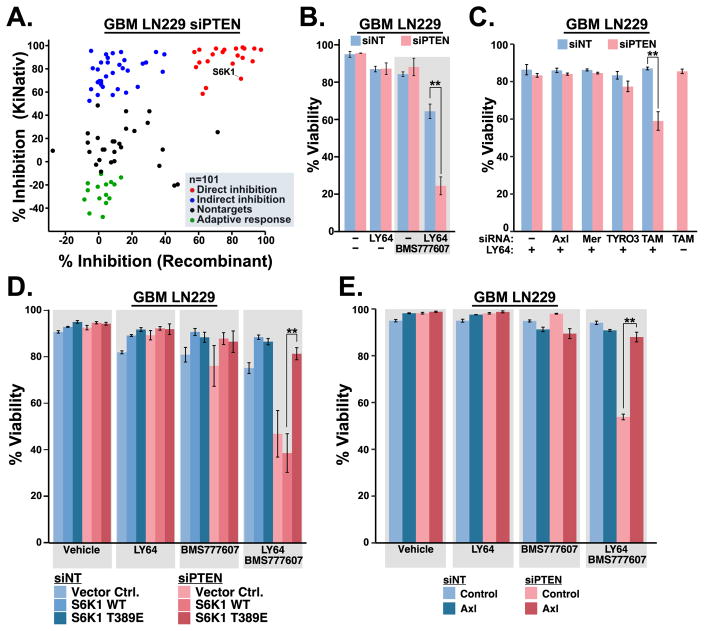
Beyond its direct effects on S6K1 signaling, we considered that the multikinase targeting portfolio of AD80 could contribute to its cytotoxic effects in PTEN-deficient cells. We surveyed the kinome-wide effects of AD80 in PTEN-knockdown LN229 cells using differential mass spectrometry analysis of kinase binding to biotin-acylated ATP analogs (KiNativ analysis, Table S1) (Patricelli et al., 2007). Comparison of KiNativ data with previously published in vitro recombinant kinase inhibition revealed three clusters of kinase responses to AD80 (Figure 4a) (Dar et al., 2012). Kinases that were inhibited ≥ 50% both in vitro and in cells appear to be direct targets, while kinases that avoided substantial inhibition in vitro may reflect context-dependent or indirect effects. Additionally, some kinases increased ATP-analog binding activity (negative inhibition), which may be indicative of adaptive signaling in response to AD80. Ingenuity Regulator Analysis of AD80 direct and indirect targets nominated the tyrosine kinase AXL for further analysis (Table S2). AXL is a member of the TYRO3, AXL, MER (TAM) family of tyrosine kinases, which are emerging as key mediators of resistance to kinase-targeted therapeutics (Elkabets et al., 2015; Scaltriti et al., 2016). Combining the S6K1 inhibitor LY-2779964 with the AXL/TAM kinase inhibitor BMS-777607 elicited substantial cytotoxicity that was selective for PTEN-deficient cells similar to AD80 (Figure 4b, S4a) (Schroeder et al., 2009). We tested the cytotoxic effects of TAM kinase knockdown in cells treated with LY-2779964. Knockdown of TAM kinases alone or in combination was not cytotoxic (Figure 4c). Nevertheless, knockdown of TAM kinases in combination with PTEN revealed a specific vulnerability to single agent LY-2779964 (Figure 4c, S4b). Expression of activated S6K1 T389E or exogenous AXL prevented cytotoxicity of combination LY-2779964 with BMS-777607, indicating the requirement for combined inactivation of S6K1 with Axl for PTEN-selective cytotoxicity (Figures 4d–e, S4c–e). Altogether the results reveal a PTEN-selective targeting strategy featuring inhibitors of S6K1 and TAM family kinases.
FIGURE 4.
S6K1/TAM kinase combination targeting. (a) siPTEN LN229 cells were cultured for 3 hours in vehicle control or 10 μM AD80, then submitted for ATP-binding site occupancy analysis (KiNativ analysis, ActivX). The % inhibition by AD80 was matched and plotted with previously published activity using recombinant enzyme assays (Dar et al., 2012). Kinase responses to AD80 were classified as indicated. (b) PTEN-selective cytotoxic effects of 5 μM LY-2779964 +/− 10 μM BMS-777607, n=3 (c) Knockdown of TAM kinases sensitizes PTEN-deficient cells to 5 μM LY-2779964, n=3. (d) Activated S6K1 T389E but not S6K1 WT mediates resistance to 5 μM LY-2779964 + 10 μM BMS-777607, n=3. (e) AXL expression mediates resistance of PTEN-deficient cells to 5 μM LY-2779964 + 10 μM BMS-777607, n=3. Mean ±SD viability was measured after 72 hours of culture; ** indicates p<0.01 by two tailed t-test (b–e). See also Figure S4.
DISCUSSION
The identification of AD80 as a multikinase inhibitor with selective cytotoxicity for PTEN-deficient cells established S6K1 as part of a kinase targeting portfolio for in vitro and in vivo cytotoxic therapy of PTEN-deficient cancers (Figure 1). Biochemical properties that distinguish AD80 from other S6K1 inhibitors are the avoidance of inhibitor-induced S6K1 phosphorylation (Figure 2d), the durable inhibition of pathway signaling (Figure 2g), and suppression of protein synthesis (Figure 3). Using AD80 as a template, we identified that the combination of S6K1-targeting LY-2779964 with TAM kinase-targeting BMS-777607 is a PTEN-selective cytotoxic therapy (Figure 4b). Importantly, the TAM kinase AXL has been shown in breast cancer to mediate resistance to rapamycin, and AXL overexpression in glioblastoma correlates with poor outcome (Elkabets et al., 2015; Hutterer et al., 2008). Single agent clinical trials with LY-2779964 have reported good pharmacokinetic properties (Hollebecque et al., 2014; Tolcher et al., 2014). BMS-777607 (also designated Aslan002) is currently in phase I clinical trial for advanced and metastatic solid tumors (NCT01721148). Altogether these results provide a rationale for the investigation and translation of these and similar agents for chemotherapeutic inhibition of S6K1 in combination with TAM kinases.
EXPERIMENTAL PROCEDURES
Cell culture
LN229, A172, U87MG, CCRF-CEM, and MOLT3 cells were purchased from ATCC; GAMG was purchased from DKMZ. S6K1−/− MEFs were the kind gift of Drs. George Thomas and Sara Kozma. TSC1−/− and TSC2−/− MEFs were the kind gift of Dr. David Kwiatkowski. 293T cells were provided by Dr. Yoon. WT-MEFs and FL5.12 cells were derived from laboratory stocks. Cells were periodically tested for mycoplasma and human lines were authenticated using fingerprint analyses from Genetica, Inc.
AD80 therapy for Pten-deficient leukemia
Donor bone marrow from PIPC-injected Mx1-Cre+;PTENfl/fl CD45.2+ B6 mice was transplanted into CD45.1 BoyJ recipient mice. Four days post-transplant, mice were randomly selected for groups treated daily for 10 days i.p. with vehicle control or 20 mg per kg of body weight AD80. Mice were monitored and euthanized upon appearance of leukemic symptoms.
Inhibitors
LY-2779964 was provided by Eli Lilly Co, or purchased from Selleck Chem (LY-2584702 tosylate, #S7704). AD80 was provided by Dr. Kevan Shokat or obtained from Cayman Chemical. DG2 was from Sigma Chemical, PF-4708671 was from Cayman Chemical, rapamycin was from LC Laboratories. Unless otherwise indicated, concentrations of inhibitors used are: 5 μM LY-2779964, 10 μM AD80, 10 μM BMS-777607.
Viability measurements
Viability of adherent cell lines was measured in triplicate wells using the CytoTox Glo assay (Promega). Viability in suspension lines was measured in technical replicates by propidium iodide exclusion in a flow cytometer.
KiNativ analysis
siPTEN LN229 cells were treated with 10 μM AD80 for 3 hours. Cells were pelleted and flash frozen before submission for KiNativ analysis (ActivX). Statistically significant results from unambiguous kinases were plotted against data from an analysis of AD80 activity using recombinant kinases (Dar et al., 2012). Kinases were assigned into four groups: ≥50 inhibition in both assays; ≥50% inhibition (KiNativ) and ≤50% (recombinant); ≤−10% inhibition (KiNativ) and ≤50% (recombinant); all others.
Translation analysis
For polysome profiles, cell extracts were loaded onto 0.5 M–1.5 M sucrose density gradients and centrifuged at 36,000 rpm for 2 hour at 4°C. Gradients were fractionated and optical density at 254 nm was continuously recorded. For 3H-leucine incorporation, cells were labeled with 10 μCi [4,5]-3H-leucine for 2 hours. Proteins precipitated from 10% trichloroacetic acid lysates were washed then solubilized in NaOH for scintillation counting.
Supplementary Material
Figure S1, related to Figure 1: Cytotoxic efficacy of S6K1 inhibitors. (a) GAMG glioblastoma cells were transfected with siNT or siPTEN, then cultured in vehicle control, 5 μM LY-2779964, or 10 μM AD80. Mean ±SD viability was determined by CytoTox Glo assay in triplicate after 72 hours of culture (top); immunoblot analysis was performed in parallel wells after 3 hours of culture (bottom). *, p<0.05 by two-tailed t-test; n=3. (b) Vector control MEF cells were treated with Vehicle control, 10 μg/ml cycloheximide (CHX), 30 ng/ml TNFα, or CHX + TNFα for 22 hours. Mean viability measured from triplicate wells were by Cytotox Glo +/− standard deviation. n=2. (c) Stably transduced shNT or shPTEN MEFs were cultured with vehicle control, 30ng/ml TNFα, 10 μMAD80, or TNFα +AD80 as shown for 8 hours. Viability was measured as in (a), n=3. (d) TSC1 or TSC2 knockout MEFs, or their respective WT counterparts, were treated with TNFα +AD80 as in (b). Viability was measured as in (a), n=3.
Figure S2, related to Figure 1f: AD80 in vivo therapy. AD80 was injected i.p. at 20 mg/kg. (a) Six hours after injection, spleen lysates were prepared for rpS6 immunoblots. (b) A single cell suspension of bone marrow cells was tested for rpS6 phosphorylation by intracellular staining and flow cytometry. n=3. (c) Mean +/− SD of body weight during AD80 therapy. There was no statistical difference in body weight during among the groups during the course of treatment (two-tailed t test).
Figure S3, related to Figure 2. S6K1 pathway dynamics. (a) IL-3 dependent FL5.12 cells stably transduced with shNT or shPTEN were cultured in the absence of IL-3 (GF). shPTEN conferred a survival advantage. (b) Comparison of 1 μM LY-2779964 and 4 μM AD80 effects on regulatory phosphorylation of S6K1 in FL5.12 shNT cells. The intensity of pT389 increase is not mirrored in pS371 or pT229. n=2. (c) LN229 cells were transfected with non-targeting (−) or raptor-targeting siRNA prior to treatment with 5 μM Ly-2779964. siRaptor reduced S6K1 T389 phosphorylation. n=3.
Figure S4, related to Figure 4. S6K1/TAM kinase combination targeting. (a) rpS6 phosphorylation in siNT or siPTEN LN229 cells treated with vehicle control, 5 μM LY-2779964, and/or 10 μM BMS-777607 for 3 hours. (b) TAM kinase knockdown LN229 cells were cultured in conditions as indicated for 3 hours. Left, Immunoblots confirm protein knockdown and LY64 efficacy. MER was not detecable. Right, Fold changes in TAM mRNA abundance, referenced to siNon-Targeting control. PTEN-knockdown induced TYRO3 andAxl mRNA, but this is not reflected in total protein levels. (c) Expression of S6K1 WT and T389E in LN229 cells. (d) LN229 cells from (c) were treated for 3 hours as indicated prior to analysis. The ratio of pS235/6:rpS6 signal, standardized to Vector/vehicle control is shown. n=2. (e) LN229 cells transduced with vector control orAxl expression constructs were treated for 3 hours as indicated. Axl expression sustained S6 phosphorylation in PTEN-deficient cells.
Table S1, related to Figure 4a. KiNativ analysis of AD80 inhibition in PTEN-deficient LN229 cells. Cells were treated with 10 uM AD80 for 3 hours prior to shipment of flash frozen packed cell pellet to ActivX for KiNativ screening.
Table S2, related to Figure 4a. Ingenuity Regulator Analysis results. Kinases identified as inhibited by AD80 in the KiNativ and Dar et al. datasets (Figure 4a) were submitted for Ingenuity Regulator Analysis.
Acknowledgments
This manuscript is dedicated to the memory of Rev. Dr. L.P. Jones, an inspiration for our work. Work was supported by 2013FZ143, 81571549, 2016BC004 and 2016-12M3026 to H.L.; R21NS100077, R01NS089815 to A.T.S.; RSG-08-293-01-CCG from the American Cancer Society, R01 CA133164, R01 CA168815, the University of Cincinnati Brain Tumor Center, and the Anna and Harold W. Huffman Endowed Chair for Glioblastoma Experimental Therapeutics to D.R.P. T.T.J. is supported by the University of Cincinnati Medical Scientist Training Program. S.K. was supported, in part, by Kanae Foundation. We acknowledge technical contributions from Ms. Kristin Bradford. We gratefully acknowledge Drs. Alex Warkentin, Kevan Shokat (UCSF), and Craig Thomas (NIH) for reagents and advice. We acknowledge Dr. Sandaruwan Geeganage (Eli Lilly and Company) for providing LY-2779964. We acknowledge Drs. Jun-Lin Guan, Maria Czyzyk-Krzeska, Tom Cunningham (University of Cincinnati) and Dan Starczynowski (Cincinnati Children’s Hospital Medical Center) for manuscript critiques.
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
H.L, X.F., K.N.E. are co-first authors contributing experimental design, results, and interpretation. C.A.B., P.S., T.T.J., Y.S. contributed experimental results and suggestions. S.K., L.N., H.E.T., S.O.Y., A.S. contributed reagents, experimental design and interpretation. D.R.P. contributed experimental design, results, interpretation and manuscript preparation.
References
- Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar AC, Das TK, Shokat KM, Cagan RL. Chemical genetic discovery of targets and anti-targets for cancer polypharmacology. Nature. 2012;486:80–84. doi: 10.1038/nature11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- Elkabets M, Pazarentzos E, Juric D, Sheng Q, Pelossof RA, Brook S, Benzaken AO, Rodon J, Morse N, Yan JJ, et al. AXL mediates resistance to PI3Kalpha inhibition by activating the EGFR/PKC/mTOR axis in head and neck and esophageal squamous cell carcinomas. Cancer Cell. 2015;27:533–546. doi: 10.1016/j.ccell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller WJ, Jackson TJ, Knight JR, Ridgway RA, Jamieson T, Karim SA, Jones C, Radulescu S, Huels DJ, Myant KB, et al. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature. 2015;517:497–500. doi: 10.1038/nature13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber Filbin M, Dabral SK, Pazyra-Murphy MF, Ramkissoon S, Kung AL, Pak E, Chung J, Theisen MA, Sun Y, Franchetti Y, et al. Coordinate activation of Shh and PI3K signaling in PTEN-deficient glioblastoma: new therapeutic opportunities. Nat Med. 2013;19:1518–1523. doi: 10.1038/nm.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez A, Sanda T, Grebliunaite R, Carracedo A, Salmena L, Ahn Y, Dahlberg S, Neuberg D, Moreau LA, Winter SS, et al. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 2009;114:647–650. doi: 10.1182/blood-2009-02-206722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollebecque A, Houede N, Cohen EE, Massard C, Italiano A, Westwood P, Bumgardner W, Miller J, Brail LH, Benhadji KA, et al. A phase Ib trial of LY2584702 tosylate, a p70 S6 inhibitor, in combination with erlotinib or everolimus in patients with solid tumours. Eur J Cancer. 2014;50:876–884. doi: 10.1016/j.ejca.2013.12.006. [DOI] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, Meyuhas O, Shokat KM, Ruggero D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutterer M, Knyazev P, Abate A, Reschke M, Maier H, Stefanova N, Knyazeva T, Barbieri V, Reindl M, Muigg A, et al. Axl and growth arrest-specific gene 6 are frequently overexpressed in human gliomas and predict poor prognosis in patients with glioblastoma multiforme. Clin Cancer Res. 2008;14:130–138. doi: 10.1158/1078-0432.CCR-07-0862. [DOI] [PubMed] [Google Scholar]
- Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardella C, Lunardi A, Fedele G, Clohessy JG, Alimonti A, Kozma SC, Thomas G, Loda M, Pandolfi PP. Differential expression of S6K2 dictates tissue-specific requirement for S6K1 in mediating aberrant mTORC1 signaling and tumorigenesis. Cancer Res. 2011;71:3669–3675. doi: 10.1158/0008-5472.CAN-10-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuzumi T, Fiedler D, Zhang C, Gray DC, Aizenstein B, Hoffman R, Shokat KM. Inhibitor hijacking of Akt activation. Nat Chem Biol. 2009;5:484–493. doi: 10.1038/nchembio.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, Aban A, Chun D, Tanner S, Kozarich JW. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry (Mosc) 2007;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- Pearce LR, Alton GR, Richter DT, Kath JC, Lingardo L, Chapman J, Hwang C, Alessi DR. Characterization of PF-4708671, a novel and highly specific inhibitor of p70 ribosomal S6 kinase (S6K1) Biochem J. 2010;431:245–255. doi: 10.1042/BJ20101024. [DOI] [PubMed] [Google Scholar]
- Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 2004;23:1761–1769. doi: 10.1038/sj.emboj.7600193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- Scaltriti M, Elkabets M, Baselga J. Molecular Pathways: AXL, a Membrane Receptor Mediator of Resistance to Therapy. Clin Cancer Res. 2016;22:1313–1317. doi: 10.1158/1078-0432.CCR-15-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder GM, An Y, Cai ZW, Chen XT, Clark C, Cornelius LA, Dai J, Gullo-Brown J, Gupta A, Henley B, et al. Discovery of N-(4-(2-amino-3-chloropyridin-4-yloxy)-3-fluorophenyl)-4-ethoxy-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide (BMS-777607), a selective and orally efficacious inhibitor of the Met kinase superfamily. J Med Chem. 2009;52:1251–1254. doi: 10.1021/jm801586s. [DOI] [PubMed] [Google Scholar]
- Shahbazian D, Parsyan A, Petroulakis E, Topisirovic I, Martineau Y, Gibbs BF, Svitkin Y, Sonenberg N. Control of cell survival and proliferation by mammalian eukaryotic initiation factor 4B. Mol Cell Biol. 2010;30:1478–1485. doi: 10.1128/MCB.01218-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- Tandon P, Gallo CA, Khatri S, Barger JF, Yepiskoposyan H, Plas DR. Requirement for ribosomal protein S6 kinase 1 to mediate glycolysis and apoptosis resistance induced by Pten deficiency. Proc Natl Acad Sci U S A. 2011;108:2361–2365. doi: 10.1073/pnas.1013629108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolcher A, Goldman J, Patnaik A, Papadopoulos KP, Westwood P, Kelly CS, Bumgardner W, Sams L, Geeganage S, Wang T, et al. A phase I trial of LY2584702 tosylate, a p70 S6 kinase inhibitor, in patients with advanced solid tumours. Eur J Cancer. 2014;50:867–875. doi: 10.1016/j.ejca.2013.11.039. [DOI] [PubMed] [Google Scholar]
- Wang BT, Ducker GS, Barczak AJ, Barbeau R, Erle DJ, Shokat KM. The mammalian target of rapamycin regulates cholesterol biosynthetic gene expression and exhibits a rapamycin-resistant transcriptional profile. Proc Natl Acad Sci U S A. 2011;108:15201–15206. doi: 10.1073/pnas.1103746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedemann LM, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figure 1: Cytotoxic efficacy of S6K1 inhibitors. (a) GAMG glioblastoma cells were transfected with siNT or siPTEN, then cultured in vehicle control, 5 μM LY-2779964, or 10 μM AD80. Mean ±SD viability was determined by CytoTox Glo assay in triplicate after 72 hours of culture (top); immunoblot analysis was performed in parallel wells after 3 hours of culture (bottom). *, p<0.05 by two-tailed t-test; n=3. (b) Vector control MEF cells were treated with Vehicle control, 10 μg/ml cycloheximide (CHX), 30 ng/ml TNFα, or CHX + TNFα for 22 hours. Mean viability measured from triplicate wells were by Cytotox Glo +/− standard deviation. n=2. (c) Stably transduced shNT or shPTEN MEFs were cultured with vehicle control, 30ng/ml TNFα, 10 μMAD80, or TNFα +AD80 as shown for 8 hours. Viability was measured as in (a), n=3. (d) TSC1 or TSC2 knockout MEFs, or their respective WT counterparts, were treated with TNFα +AD80 as in (b). Viability was measured as in (a), n=3.
Figure S2, related to Figure 1f: AD80 in vivo therapy. AD80 was injected i.p. at 20 mg/kg. (a) Six hours after injection, spleen lysates were prepared for rpS6 immunoblots. (b) A single cell suspension of bone marrow cells was tested for rpS6 phosphorylation by intracellular staining and flow cytometry. n=3. (c) Mean +/− SD of body weight during AD80 therapy. There was no statistical difference in body weight during among the groups during the course of treatment (two-tailed t test).
Figure S3, related to Figure 2. S6K1 pathway dynamics. (a) IL-3 dependent FL5.12 cells stably transduced with shNT or shPTEN were cultured in the absence of IL-3 (GF). shPTEN conferred a survival advantage. (b) Comparison of 1 μM LY-2779964 and 4 μM AD80 effects on regulatory phosphorylation of S6K1 in FL5.12 shNT cells. The intensity of pT389 increase is not mirrored in pS371 or pT229. n=2. (c) LN229 cells were transfected with non-targeting (−) or raptor-targeting siRNA prior to treatment with 5 μM Ly-2779964. siRaptor reduced S6K1 T389 phosphorylation. n=3.
Figure S4, related to Figure 4. S6K1/TAM kinase combination targeting. (a) rpS6 phosphorylation in siNT or siPTEN LN229 cells treated with vehicle control, 5 μM LY-2779964, and/or 10 μM BMS-777607 for 3 hours. (b) TAM kinase knockdown LN229 cells were cultured in conditions as indicated for 3 hours. Left, Immunoblots confirm protein knockdown and LY64 efficacy. MER was not detecable. Right, Fold changes in TAM mRNA abundance, referenced to siNon-Targeting control. PTEN-knockdown induced TYRO3 andAxl mRNA, but this is not reflected in total protein levels. (c) Expression of S6K1 WT and T389E in LN229 cells. (d) LN229 cells from (c) were treated for 3 hours as indicated prior to analysis. The ratio of pS235/6:rpS6 signal, standardized to Vector/vehicle control is shown. n=2. (e) LN229 cells transduced with vector control orAxl expression constructs were treated for 3 hours as indicated. Axl expression sustained S6 phosphorylation in PTEN-deficient cells.
Table S1, related to Figure 4a. KiNativ analysis of AD80 inhibition in PTEN-deficient LN229 cells. Cells were treated with 10 uM AD80 for 3 hours prior to shipment of flash frozen packed cell pellet to ActivX for KiNativ screening.
Table S2, related to Figure 4a. Ingenuity Regulator Analysis results. Kinases identified as inhibited by AD80 in the KiNativ and Dar et al. datasets (Figure 4a) were submitted for Ingenuity Regulator Analysis.