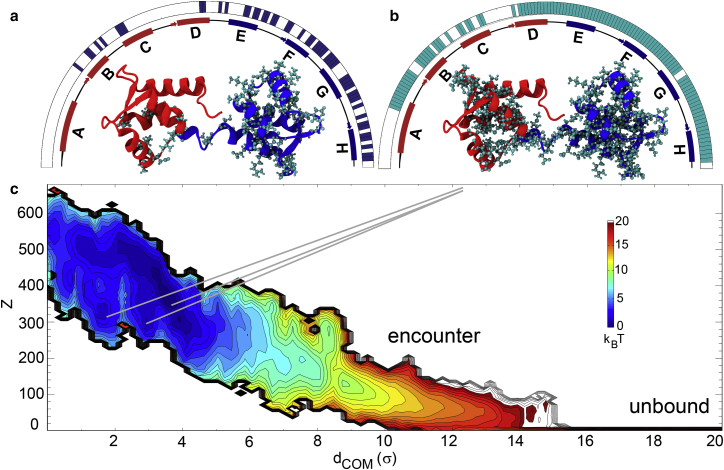
Figure 2.
Reweighted PMF of apoCaM-Ng13–49 binding and changes in chemical shifts for apoCaM upon Ng13–49 binding. (a and b) The residues that experienced significant changes in chemical shifts are projected on the apoCaM structure (PDB: 1CFD) as a ball-and-stick representation from experimental measurement in (a) (30) and from our calculations according to the coarse-grained molecular simulations in (b). (c) The PMF from the coarse-grained molecular simulations is plotted against the center of mass distance between apoCaM and Ng13–49 (dCOM) and the number of intermolecular contacts (Z). The coarse-grained molecular simulations were performed at pH = 6.3 and ionic strength = 0.1 M according to the conditions of the nuclear magnetic resonance experiments (30). We used a reduced unit of length σ = 3.8Å. The color is scaled in kBT, T = 1.1 ε/kB, where ε is reduced unit of energy and kB is Boltzmann constant. For visual guidance, we provided bars above the schematic representation of secondary structures in red and blue segments of apoCaM on the half circles in (a and b). To see this figure in color, go online.