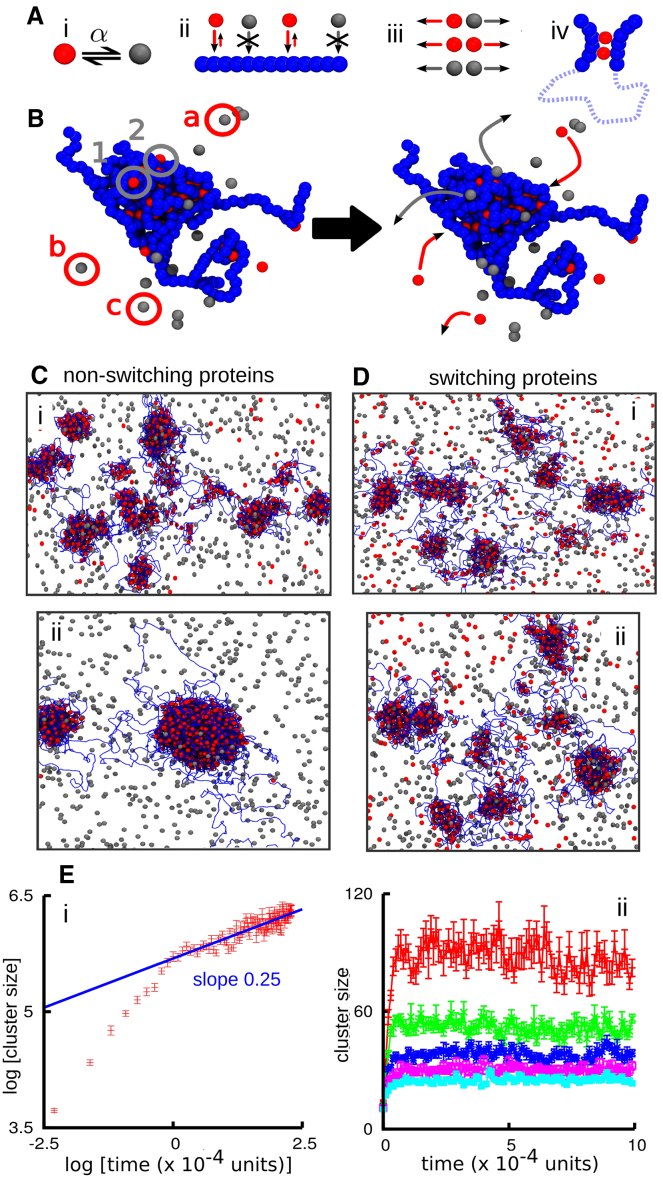
Figure 1.
Protein switching arrests cluster coarsening. In (A)–(D), active and inactive proteins are colored red and gray, respectively; chromatin is represented by strings of blue beads. (A) Schematic of the model (Brownian dynamics simulations). (i) Proteins (single spheres) switch between red and gray states at rate α. (ii) Only proteins in the red state can bind chromatin. (iii) Red and gray beads interact via steric repulsion only. (iv) Proteins can bind to ≥2 sites to create molecular bridges and loops. (B) Snapshots illustrating protein binding/unbinding. Bound active proteins have clustered and compacted chromatin. Bound active proteins 1 and 2 (gray circles) switch and become inactive and dissociate (gray arrows); inactive proteins a–c in the soluble pool (red circles) are activated and may bind to the cluster (red arrows). (C) Snapshots taken (i) 104 and (ii) 2 × 104 simulation units after equilibration. The simulation involved a 5000-bead fiber (corresponding to 15 Mbp) and N = 4000 nonswitchable proteins, of which half are able to bind. (D) As in (C), but for N = 4000 switchable proteins (α = 0.0003 inverse Brownian times). (E) Average cluster size as a function of time. Error bars denote standard deviations of the mean. (i) Nonswitching proteins. (ii) Switching proteins; from top to bottom, α equals 0.0001, 0.0002, 0.0003, 0.0004, and 0.0005 inverse Brownian times (or α−1 ≃ 10–60 s in real units). To see this figure in color, go online.