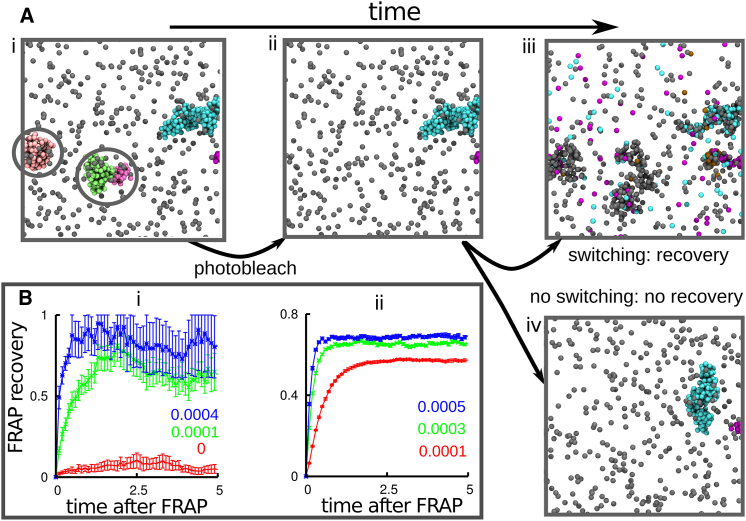
Figure 3.
In silico FRAP (Brownian dynamics simulations). (A) Snapshots taken 104 (i and ii) or 2 × 104 (iii and iv) after equilibration, during an in silico FRAP experiment (only proteins—and not chromatin beads—are shown for clarity). (i) The simulation begins with N = 2000 equilibrium proteins, half of which are able to bind the chromatin fiber, both specifically (interaction strength 15 kBT, cutoff 1.8σ) to every 20th bead in the polymer, and nonspecifically (interaction strength 4 kBT, cutoff 1.8σ) to any other bead. After 104 time units, a structure with multiple clusters forms. The snapshot shows only a portion of this, for clarity; five clusters of bound proteins have developed (unbound proteins are gray; bound proteins in the five clusters are blue, pink, purple, and green). Circled areas will be photobleached. (ii) Photobleaching involves making bound proteins invisible (the bleached proteins are still present in the simulation). (iii) If proteins can switch, clusters reappear in the same general place (as new proteins replace their bleached counterparts). (iv) If proteins cannot switch (i.e., α = 0), clusters do not recover (as their protein constituents do not recycle). (B) FRAP recovery. Error bars give SD of mean, and time is given in multiples of 104 simulation units; the values of α, in units of inverse Brownian times, are as indicated in each panel. Only the postbleaching signal is shown (the prebleaching value would be constant and equal to 1 in these units). (i) Number of unbleached proteins in the bleached volume (a sphere of 50σ) as a function of time, after bleaching. The signal is normalized with respect to the number of proteins initially in the bleached volume. (ii) Number of unbleached proteins in clusters as a function of time after bleaching, after all proteins in clusters at a given instant are bleached. The signal is normalized with respect to the proteins in clusters at the time of bleaching. To see this figure in color, go online.