Figure 1.
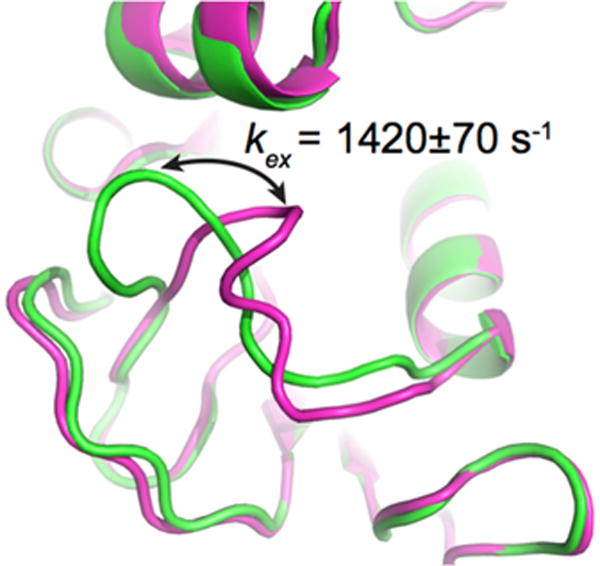
Conformational exchange experienced by the Met20 loop in E. coli DHFR, as probed by 15N-CPMG NMR relaxation dispersion experiments. The CPMG experiment is particularly well suited to extract exchange rates (kex) between two or more conformations in solution on the timescale of catalysis (ms) in many enzyme systems, offering a measure of comparison between conformational exchange experienced by the enzyme and its catalytic rate (kcat). This method can also provide quantitative information on low-populated “invisible” excited sub-states and theft populations in solution (pA and pB). The cartoon representation illustrates a simplified view of the closed (green) and occluded (magenta) conformations sampled by the Met20 loop in E. coli DHFR as it catalyzes hydride transfer, an atomic-scale movement essential to bacterial DHFR function and correlated with substrate/cofactor recognition and turnover in this enzyme (reviewed in [28,35,40]). Depicted PDB structures are 1RX2 (green) and 1RX7 (magenta).
