Figure 5.
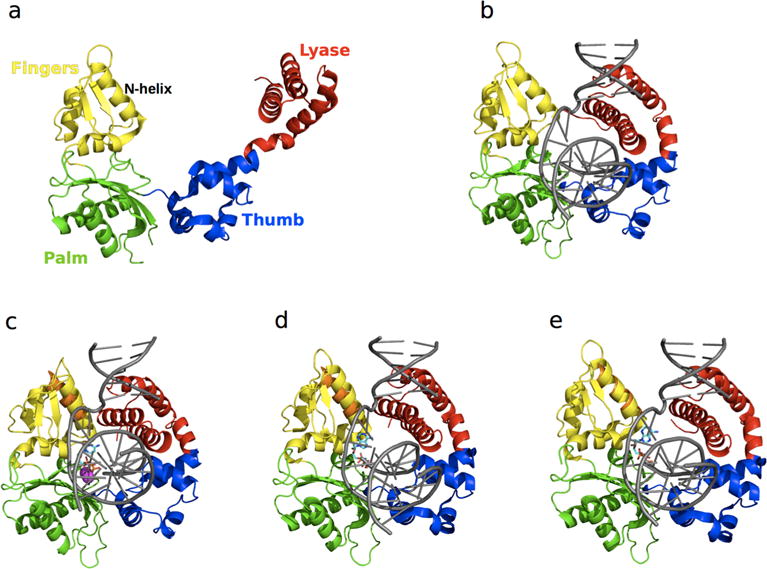
Structure of free and ligand-bound states of Pol β. The lyase domain is shown in red while the thumb, palm, and fingers subdomains of the polymerase domain are shown in blue, green and yellow, respectively. Pol β in the (a) apo (PDB 1BPD) form is characterized by an open conformation; (b) binary complex (PDB 1BPX) formation is accompanied by a 40 Ǻ conformational shift of the lyase domain to form a compact “open” conformation; (c) matched dNTP-bound ternary (PDB 1BPY) complex shows a 10 A shift of the N-helix of the fingers domain relative to the binary form (in orange), leading to the formation of a “closed” conformation. Binding of mismatched dNTPs has been shown to adopt (d) closed (PDB3C2M) and (e) open (PDB 4F5P) conformations of the N-helix. For oomparison, the N-helix from the binary form is shown in orange in the ternary complexes in c–e, illustrating subtle yet functionally significant conformational exchange in this enzyme. DNA substrates in all the binary and ternary forms are shown in grey (b–e), the dNTPs in c–e are shown as cyan sticks, and the Mg2+ ions are represented as magenta spheres in c.
