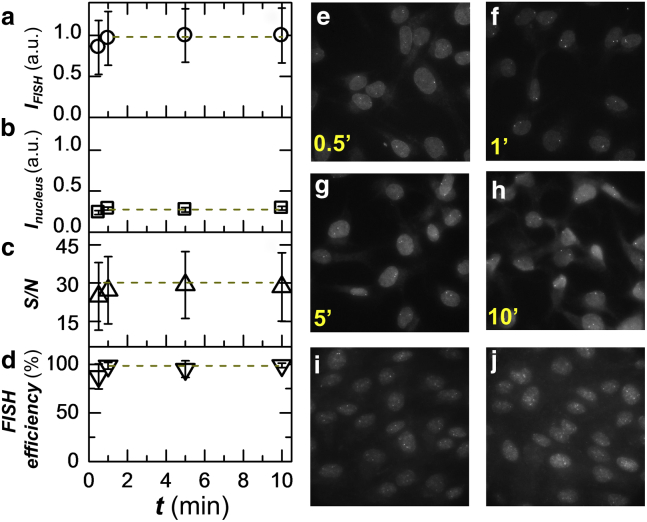
Figure 2.
Rapid staining can be achieved with oligo DNA probes under optimized DNA FISH protocol. (a) Kinetics of the rapid DNA FISH staining. Intensity of FISH puncta for a tandemly repetitive sequence in the human genome is shown. Intensity is averaged over 300+ puncta and error bars denote SD. (b) Intensity of nuclear background staining at different time points. (c) The signal-to-noise ratio of FISH puncta. (d) The efficiency of FISH staining, calculated as the ratio of observed to expected number of puncta. (e–h) Representative images at different time points of staining. Maximum intensity projection is shown for each 3D z-stack. (i–j) Representative images of two genomic regions denoted as NR1 (i) and NR2 (j) of nonrepetitive sequence labeled by tiling ∼200 probes over a 40-kb region with a staining time of 2 min. See Materials and Methods for details on probe sequence. Maximum intensity projection is shown for each 3D z-stack. To see this figure in color, go online.