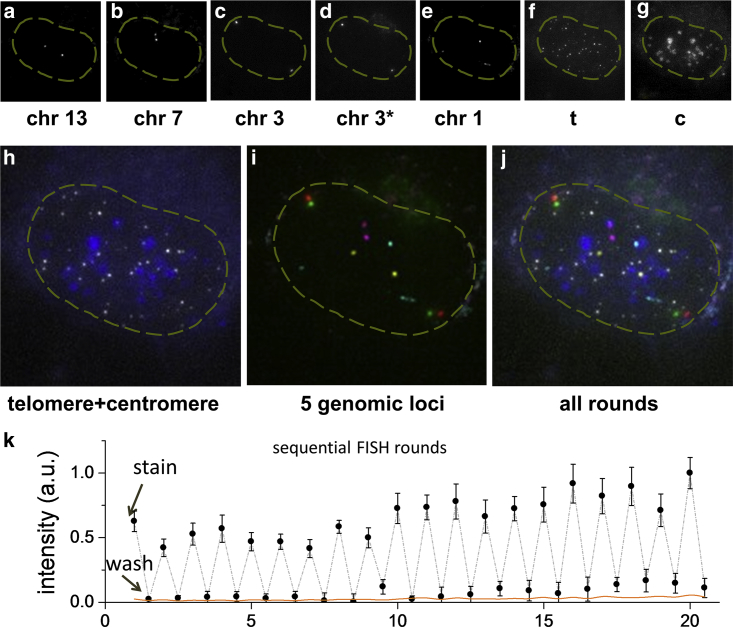
Figure 3.
Multiple genomic loci can be resolved through multiple sequential rounds of DNA FISH. (a–g) Chr13, Chr7, Chr3, Chr3∗, Chr1, telomere, and centromere are sequentially resolved in six rounds of staining of 1 min and wash of 0.5 min. Two sites 300 kb apart on chromosome 3 (denoted as Chr3 and Chr3∗) are costained and distinguished in one round with two different colors. Maximum intensity projection is shown for each 3D z-stack. (Dashed yellow line) Nucleus contour. (h) False-color overlay of telomere (white) and centromere (blue) staining. (i) False-color overlay of Chr13 (yellow), Chr7 (magenta), Chr3 (red), Chr3∗ (green), and Chr1 (cyan). (j) Overlay of all loci determined in the sequential DNA FISH. Color is the same as in (h) and (i). Image area in (a)–(i) is 22 × 22 μm. (k) Peak intensity value of 20 rounds of repeated staining of 1 min and washing of 0.5 min with nuclear background subtracted, averaged over eight loci. Error bars denote SD. The nuclear background (orange line) is calculated as the average intensity within a nuclear region minus the average intensity from the open region without cells.