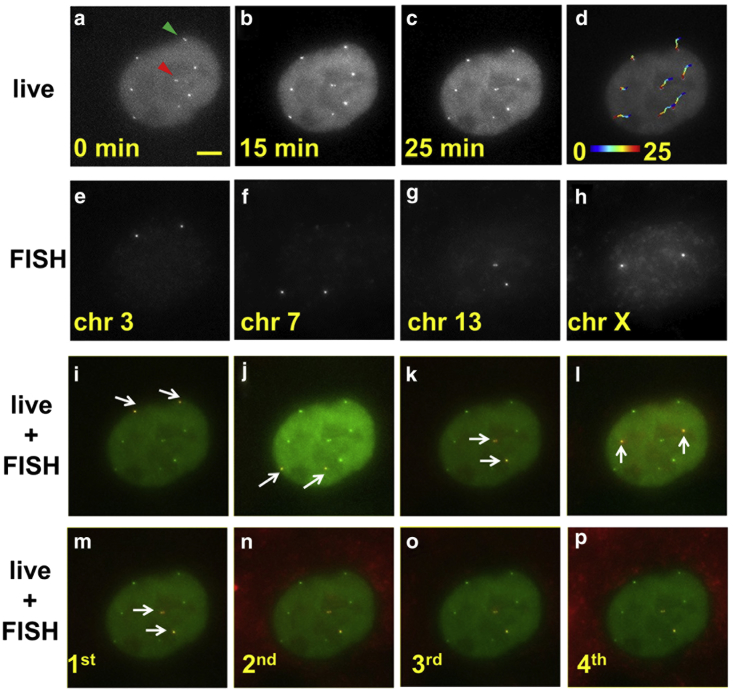
Figure 4.
Sequential rounds of DNA FISH resolve identities of loci imaged in live-cell mode. (a–c) Representative micrographs of a cell from time-lapse images. Eight spots corresponding to four genomic regions in a diploid RPE cell are seen. (Green arrowhead) Pair of sister chromatids that are distinguished at the beginning of the live imaging, and that came closer than the diffraction limit at a later time. (Red arrowhead) Another pair of sister chromatids that are distinct throughout the image acquisition with fluctuating separation. (d) Overlay of the cell image at the end of the live observation with the corresponding loci trajectories. (Color) Time in minutes. (e–h) FISH images in four sequential FISH rounds reveal the identities of the loci. (i–l) Overlay of live images at 25 min with DNA FISH images from each round shows faithful registration and negligible nuclear deformation. (White arrows) FISH spots in each round. (m–p) Four sequential DNA FISH rounds staining Chr13 show consistent image registration between rounds. All micrographs here show the same image area and scale bar in (a) as 5 μm.