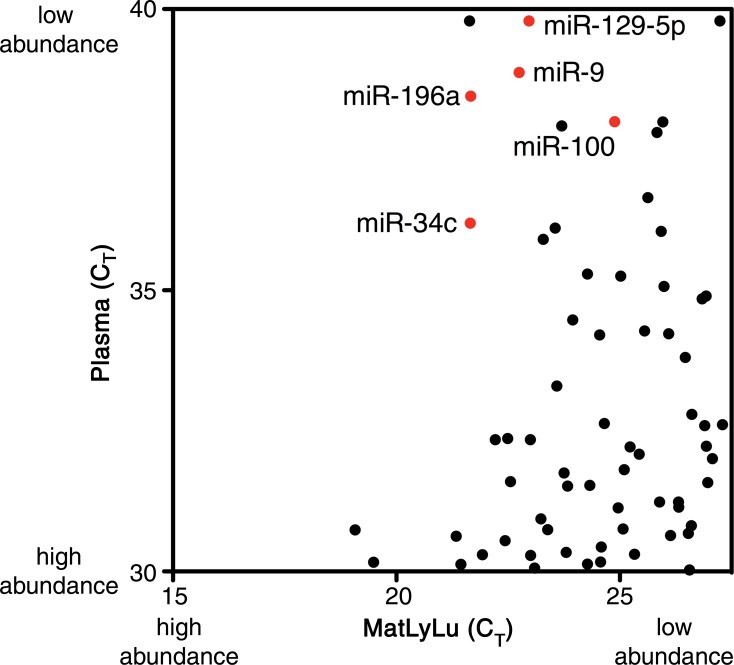
Figure 5b:
Scatterplots generated for the identification of candidate tumor biomarkers in the MatLyLu rat model system. miRNA expression profiling was performed by using RT-qPCR arrays to analyze RNA purified from the plasma of untreated control rats (n = 4) or from the MatLyLu tumor cell line. (a) miRNAs with higher abundance in the cell line (lower cycle threshold [CT] values on the x-axis) and low or undetected abundance in untreated rat plasma (higher cycle threshold values on the y-axis) were selected as candidate biomarkers (shown in red). The nonbiomarker internal control miR-16 is shown in green. (b) Higher resolution image of the upper left quadrant of a can be used to identify candidate miRNAs. Candidate biomarkers are shown in red, and other miRNAs are shown in black.