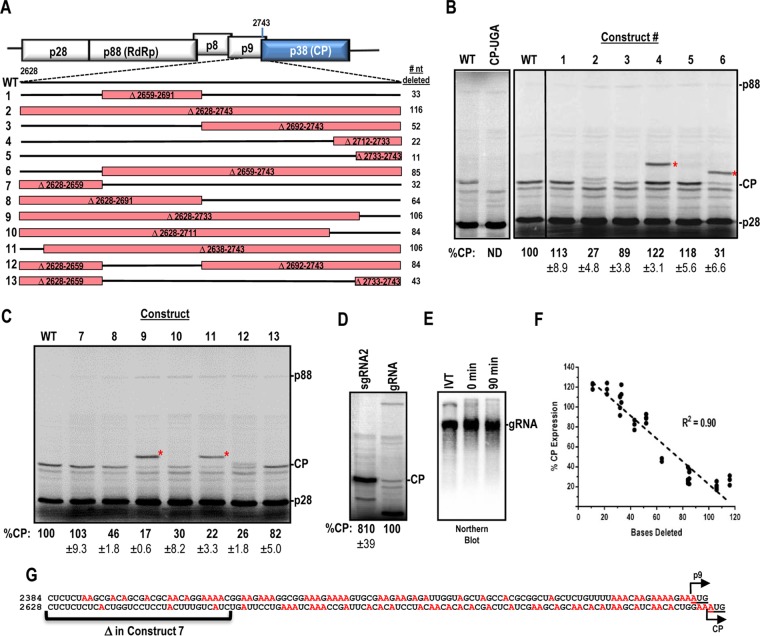
FIG 1.
Deletion analysis of a region required for internal expression of the TCV CP. (A) Deletions generated in a 116-nt region immediately upstream of the CP ORF. Deleted regions are boxed in red. Construct designations are shown to the left, and numbers of nucleotides deleted are shown to the right. (B and C) In vitro translation of constructs 1 to 13 by use of WGE. CP-UGA contains an in-frame stop codon at position 2759 in the TCV genome. Asterisks denote locations of p9-CP fusion proteins that result from internal translation initiation at the p9 ORF for some constructs. Data were normalized to p28 levels, and means and standard deviations are shown. Data are from at least three independent experiments. (D) CP expression in WGE, using equimolar amounts of sgRNA2 and gRNA. (E) Analysis of RNA integrity during the WGE assay. After 90 min of incubation in WGE, the mixture was subjected to Northern blotting using a gRNA probe. Purified RNA transcripts (IVT) and the 0-min time point were used as controls. (F) Inverse correlation between CP expression and deletion size. Normalized CP expression was plotted against the number of nucleotides deleted for each construct in panel A. Linear regression was used to determine the goodness of fit as shown by the R2 value. (G) A-rich region upstream of the p9 and CP ORFs. Adenine residues are shown in red. Initiation codons are underlined.