Fig. 2.
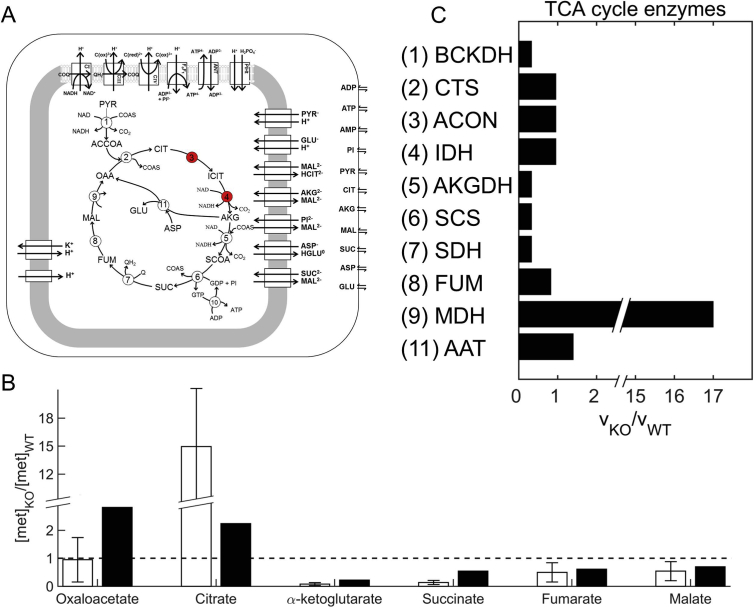
Reaction fluxes based on changes in gene transcription obtained in response to a certain stress condition are sufficient to mimic the metabolic phenotype corresponding to that condition. A) Schematic representation of the thermodynamically balanced computer model of mitochondrial oxidative phosphorylation used to predict relative changes in metabolites on the basis of relative changes in estimated reaction fluxes under wild-type (WT) and knockout (KO) conditions; figure modified and reproduced from (Wu et al., 2007). The two red circles represent the two genes that were deleted in a previous study (Ke et al., 2015). B) Alterations in the concentrations of TCA cycle metabolites after the deletion of two TCA cycle enzymes. Open bars show experimentally observed values and closed bars the computationally predicted values. Metabolite concentrations were computed by using the model shown in A and reaction flux ratios shown in C. C) Median relative changes in TCA cycle enzyme fluxes, estimated by using the approach shown in Fig. 1, during the intraerythrocytic development cycle under two conditions (WT and KO). The effect of the stress condition, i.e., knockout of two TCA cycle enzymes, is captured by the transcriptome data obtained under WT and KO conditions (Ke et al., 2015). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)