Figure 4.
NR2C Inhibition Alters Spatiotemporal Wave of Clock Gene Expression by Selectively Impairing Synchronization of Dorsal NR2C+ SCN Neurons
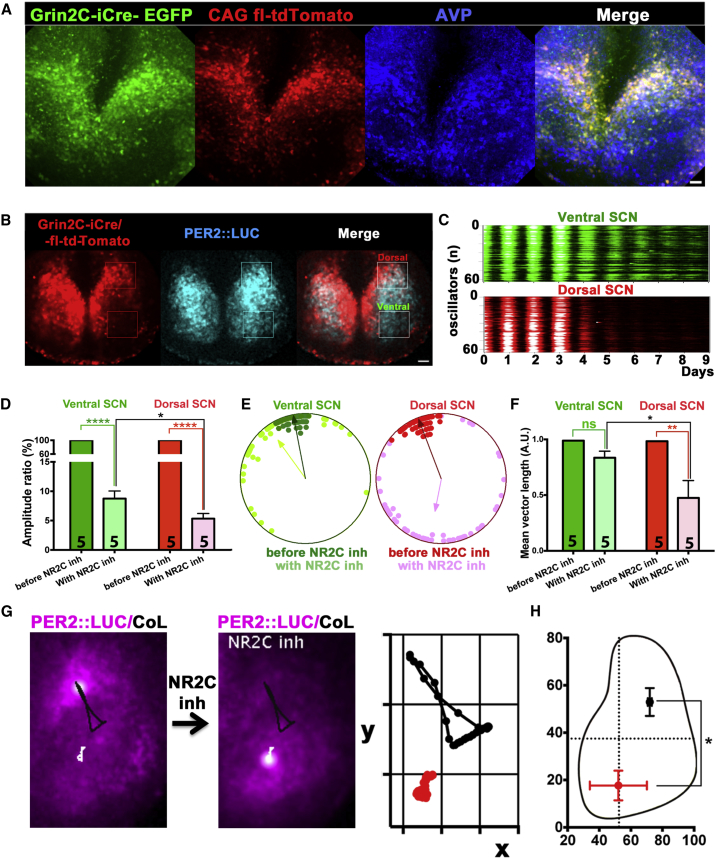
(A) Confocal micrographs of SCN slices from Grin2C-iCre mice transduced with AAV-CAG-flex-tdTomato and counterstained with AVP antiserum to highlight SCN cytoarchitecture.
(B) Representative frame from time-lapse recording of PER2::LUC SCN slices from Grin2C-iCre mice expressing flex-tdTomato to label NR2C+ neurons. Boxed areas show dorsal and ventral SCN, as defined by tdTomato signal.
(C and D) Raster plots (C) and bar graphs (D) of amplitude ratio of PER2::LUC oscillations in ventral and dorsal regions of SCN slices treated with DQP-1105.
(E and F) Representative Rayleigh plots (E) and bar graphs (F) of mean vector length of ventral and dorsal PER2::LUC oscillators, as defined by Grin2C-tdTomato expression, before DQP-1105 treatment and in the presence of the drug.
(G and H) (G) Photomicrographs and Poincaré plots of spatiotemporal waves of PER2::LUC quantified by CoL before (black trajectory) and during (white trajectory; red on Poincaré plot) DQP-1105 treatment, within a single SCN and across several replicates (H). SCN outline is for presentation purposes.
All bar graphs are mean ± SEM; n experimental replicates depicted on bars. Statistical tests are as follows: (D and F) two-way repeated-measures ANOVA, Bonferroni corrected; (G) paired two-tailed t test, n = 3. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001. Scale bars, 50 μm. See also Figure S7 and Movie S2.