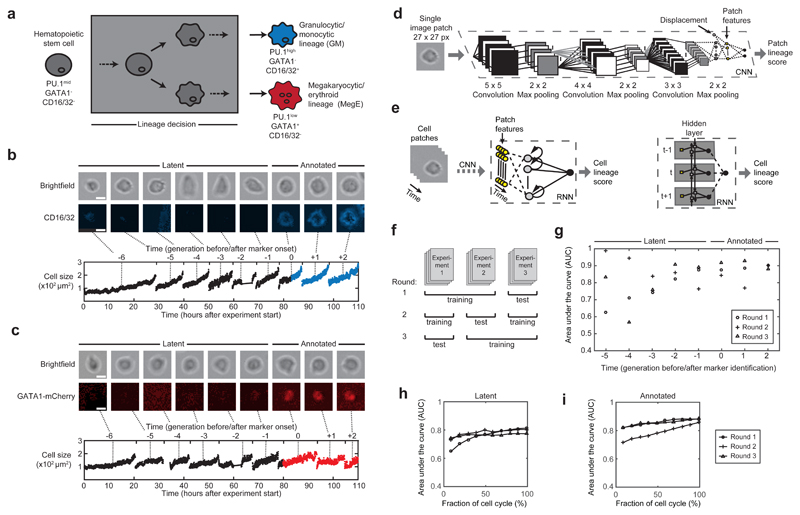
Figure 1. Prediction of hematopoietic lineage choice up to three generations before molecular marker annotation using deep neural networks.
(a) Hematopoietic stem cells (gray) can differentiate and are annotated as committed towards the granulocytic/monocytic (GM, blue) lineage via detection of CD16/32, or towards the megakaryocytic/erythroid lineage (MegE, red) via GATA1-mCherry expression. These conventional markers necessarily appear after the lineage decision (gray box). (b,c) Exemplary image patches of a branch of single cells committing to either GM (b, upper row) or MegE (c, upper row) lineage (scale bars: 10µm). Cells with no marker expression are called “latent”, cells with marker expression “annotated” (b,c, middle row). Our automatic image processing pipeline allows robust cell identification and thus quantification of movement and morphology (demonstrated with cell size in lower rows of b and c) (d) A single image patch and the according cell’s displacement (white node) with respect to the previous time point are fed into a convolutional neural network (CNN) consisting of convolutional and fully connected layers (see Methods for more details on the network architecture). The last fully connected hidden layer (yellow) can be interpreted as patch-specific features. (e) To account for temporal dependencies we feed the CNN-derived patch features of a cell (yellow) in a recurrent neural network (RNN). The nodes in the hidden layer are connected to output nodes as well as all other hidden nodes across time (left); this temporal dependency is further illustrated in an unrolled representation of the RNN (right), where yellow squares represent the patch feature vectors at a specific time point and forward/backward arrows reflect the bidirectional architecture of the RNN. Every patch is assigned a lineage score between 0 and 1 (0=MegE, 1=GM, 0.5=unsure). (f) Two experiments are used for training, while one experiment is left out to assess generalization quality of the learned model. We repeat this procedure three times in a round-robin fashion. (g) Area under the receiver operating characteristics curve (AUC; 1.0=perfect classification, 0.5=random guessing) determines the performance of the trained models. Annotated cells (generations 0,+1,+2) and latent cells up to three generations before marker onset (generations -3,-2,-1) show AUCs higher than 0.77 (n=3 rounds, 4204 single cells in total). (h,i) AUCs when only (contiguous) subsets of image patches are used to compute the cell lineage score. AUCs over 0.75 were reached when using the first ~25% of timepoints in the cell cycle from latent (h) and annotated cells (i), respectively.