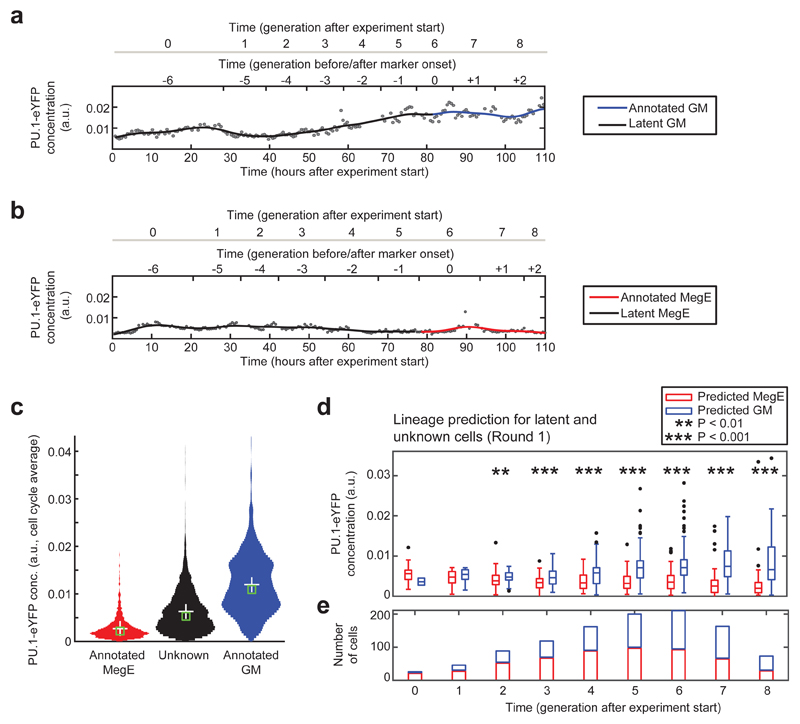
Figure 2. Subsets of cells with differential PU.1-eYFP expression can be distinguished two generations after experiment start.
(a) Increase of PU.1-eYFP concentration in a branch with annotated GM marker onset. (b) Decrease of PU.1-eYFP concentration in a branch with annotated MegE marker onset. Concentrations (black dots) are fitted with a B-spline (black/colored line). (c) Cells without marker expression (unknown or latent fate, black) show an intermediate PU.1-eYFP concentration compared to cells annotated for GM (blue) and MegE (red) lineage. (d) Concentration of PU.1-eYFP for unknown and latent cells in generations after experiment start, subdivided into predicted GM (blue) and MegE (red), based on CNN-RNN lineage score for round 1 (see Supplementary Fig. 4 for rounds 2 and 3). PU.1-eYFP concentration is significantly different in generation 2 (P<0.01) and 3-8 (P<0.001, unpaired wilcoxon rank-sum test) after experiment start between the two predicted groups. (e) Significantly different PU.1-eYFP expression in generation 2 after experiment start is detected in 55 MegE (red) vs. 34 GM (blue) predicted cells, respectively.