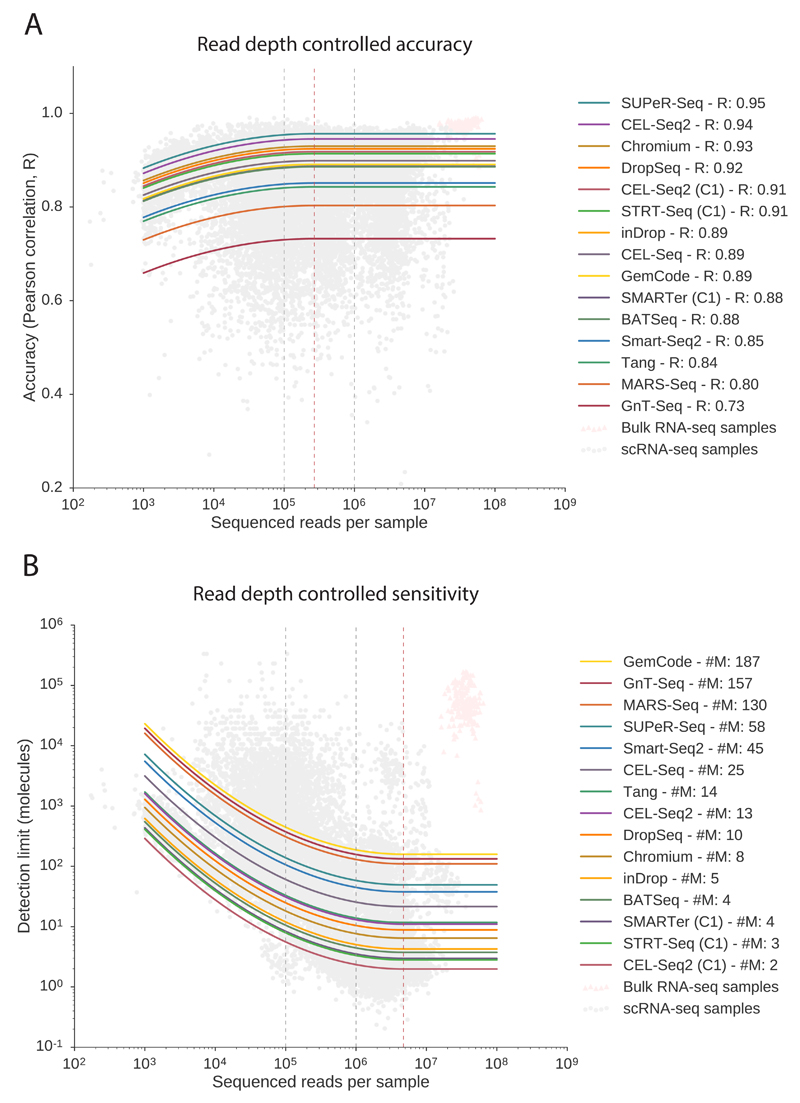
Figure 3. Performance metrics after accounting for sequencing depth.
Both accuracy and sensitivity are modeled with a global dependency on sequencing depth considering diminishing returns, with a distinct corrected performance parameter for each protocol. Both models (one for accuracy and one for sensitivity) has 26 parameters, and are fitted to n=20,717 observations (the number of samples). (Bulk data in pink triangles are only displayed for context). Solid curves show the predicted dependence on sequencing depth. (A) Accuracy is only marginally dependent on sequencing depth. Saturation occurs at 270,000 reads per cell in the model (dashed red line). Protocol names are ordered by performance based on predicted correlation R at 1 million reads. (B) Sensitivity is critically dependent on sequencing depth and accounting for read depth is key for fair comparison of protocols. Saturation identified by the model occurs at 4.6 million reads per cell (dashed red line). The gain from 1 to 4 million reads per sample is marginal, while we note that moving from 100,000 reads to 1 millions reads corresponds to an order of magnitude gain in sensitivity (dashed black lines). Protocols are ordered by performance based on predicted detection limit (#M, number of molecules at 1 million reads).