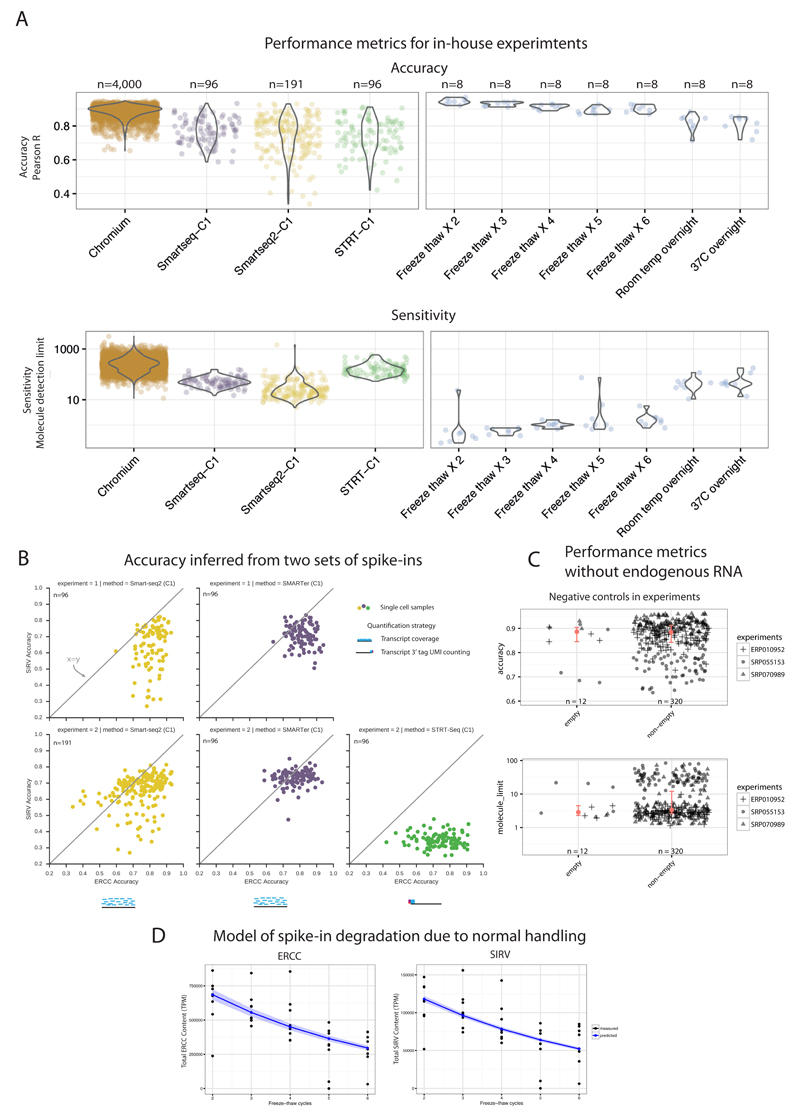
Figure 4. Investigation of factors with potential impact on performance metrics.
(A) Batch effects and RNA degradation. Performance distributions for three protocols implemented as a single batch, on the Fluidigm C1 platform (left) and on the 10x Chromium (far-left; different batch). Performance distributions of spike-ins measured after freeze-thaw cycles, normal (2-3 cycles) to critical degradation (6 cycles, left overnight at room temperature). (B) Accuracy estimates across both ERCC and SIRV spike-ins are similar. Accuracy (Pearson correlation R) of both spike-ins (ERCC’s and SIRV’s) inferred across two replicates using multiple protocols. (C) Endogenous mRNA amount does not affect performance metrics. Comparison of performance metrics between empty (lacking endogenous mRNA) and non-empty samples from 3 published datasets shows similar performance and no bias due to presence of endogenous mRNA. Red dot shows median, and red bar shows 95% confidence interval of median, estimated with bootstraps. Empty accuracy CI is 100% contained in non-empty CI, and empty sensitivity CI is 84% contained in non-empty CI. (D) Model of relative spike-in abundance degradation during normal handling. Posterior predictions from Bayesian exponential decay model, for both ERCC’s and SIRV’s. The decay parameter was 19% and 18.5%, respectively. Confidence bands correspond to 95% confidence interval from posterior parameter distribution.