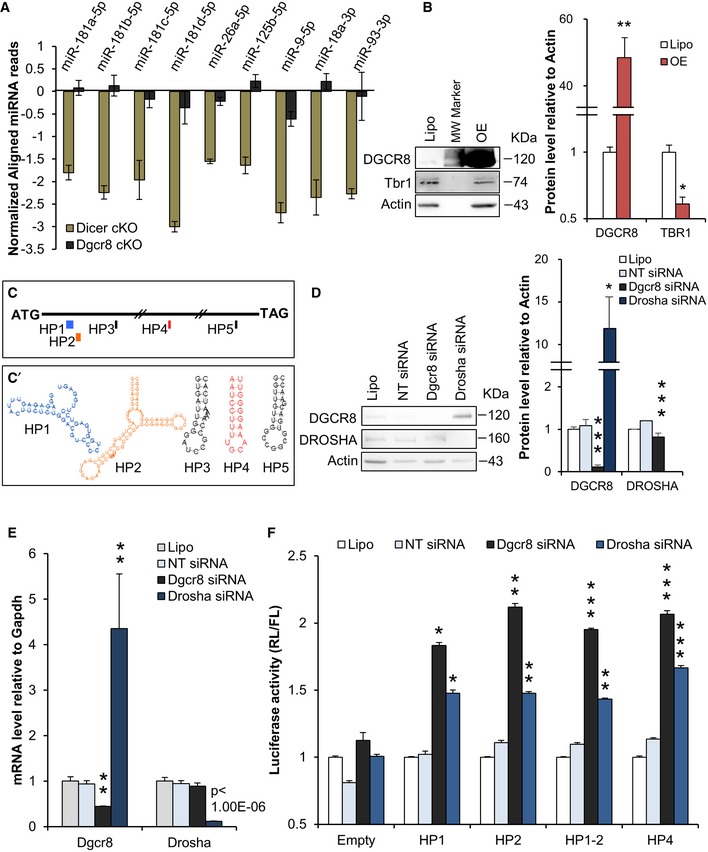
-
A
Read counts of experimentally validated miRNAs targeting the Tbr1 transcript in E13.5 Dicer cKO and Dgcr8 cKO cortices.
-
B
Western blot and quantification of DGCR8, TBR1, and actin expression in N2A cells upon DGCR8 overexpression.
-
C
Position of the 5 predicted hairpins (HPs) in Tbr1 mRNA coding sequence (ATG‐TAG, black thin segment).
-
C’
Predicted secondary structures of the 5 HPs (color code as in C).
-
D
Western blot and quantification of DGCR8, DROSHA, and actin expression in N2A cells upon Dgcr8 or Drosha knockdown.
-
E
qRT–PCR analysis of endogenous Dgcr8 and Drosha mRNA in N2A cells upon Dgcr8 or Drosha knockdown.
-
F
N2A cells transfected with Dgcr8, Drosha, or control (NT) siRNAs, respectively, co‐transfected with vectors described in (C) and (C’).
Data information: Mock (Lipo); DGCR8 overexpression (OE); non‐target (NT) siRNA. Bars indicate the variation from the mean (SEM;
n = 3 biological replicas), and
P‐values are indicated (paired Student's
t‐test); *
P < 0.05; **
P < 0.01; ***
P < 0.001.