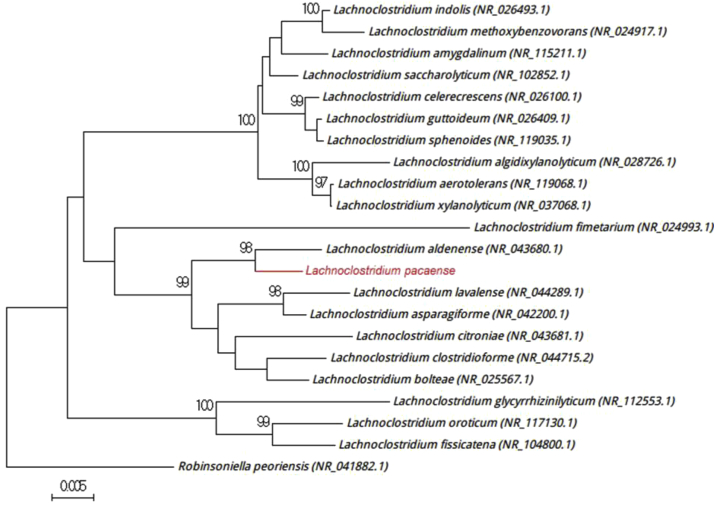
Fig. 3.
Phylogenetic tree showing position of ‘Lachnoclostridium pacaense’ Marseille-P3100T relative to other phylogenetically close neighbours. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 500 times to generate majority consensus tree. Only bootstraps scores of at least 90% were retained. Scale bar indicates 0.5% nucleotide sequence divergence.