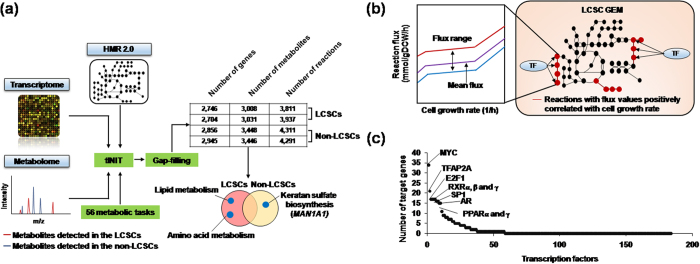
Figure 3. Reconstruction of LCSC and non-LCSC GEMs and application for the identification of regulators associated with cell proliferation.
(a) Reconstruction process of LCSC and non-LCSC GEMs. Transcriptome and metabolome data were integrated with a generic human GEM, HMR 2.027. During the GEM reconstruction process, draft GEMs were examined and validated by simulating the predefined 56 metabolic tasks under the cultivation condition of RPMI-1640 medium (Supplementary Table S5). The GrowMatch gap-filling algorithm was performed to enable the cell type-specific GEMs to demonstrate growth50. Reactions involved in lipid and amino acid metabolisms were more included in LCSC GEMs, whereas keratan sulfate biosynthetic pathway (largely mediated by MAN1A1) was more represented in non-LCSC GEMs. (b) Scheme of metabolic simulation using flux response analysis of the LCSC and non-LCSC GEMs to predict transcription factors regulating biomass generation (i.e., cell growth rate). The mean flux value (purple line) of each reaction was used when calculating the correlation between each reaction’s flux value and cell growth rate. In the right panel, upon prediction of reactions with flux values positively correlated with cell growth rate (red lines). Transcription factors (TF) that are known to bind to and regulate genes of the predicted reactions were next searched from RegNetwork31. (c) Transcription factors that potentially regulate genes responsible for the metabolic reactions with flux values positively correlated with cell growth rate. Names of the top 10 transcription factors (out of 184 transcription factors) are presented.