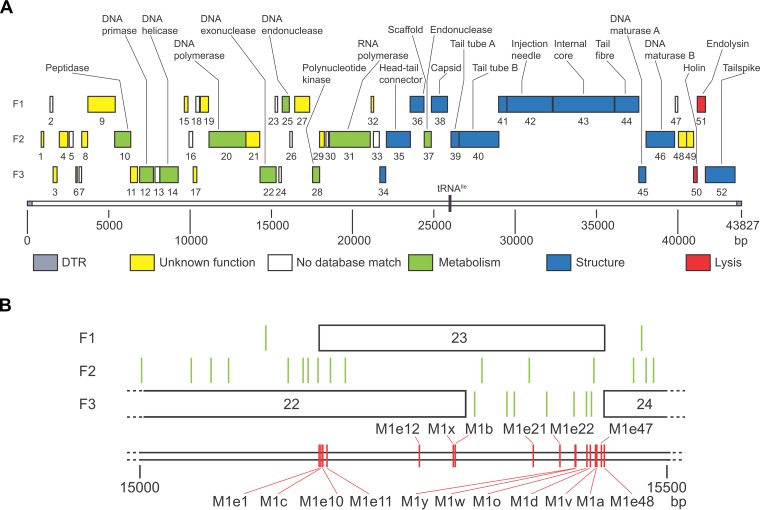
FIG 1.
Genomic map of ΦM1 wild type and its escape locus. (A) All 52 annotated ORFs are coded on the forward reading strand, in a linear progression from metabolic genes to structural genes and, finally, to host cell lysis genes. Each forward reading frame is labeled F1, F2, or F3. ORFs are shown to scale as shaded boxes numbered with the gene number, colored according to the predicted role. The single tRNAIle gene is positioned on the scale, shown in purple. Where it was possible to identify a protein by homology searches, that ORF is labeled. The scale is in base pairs. The figure was drawn to scale using Adobe Illustrator. (B) Schematic of the escape locus of ΦM1. All escape phage mutations are within phiM1-23. Each forward reading frame is labeled F1, F2, or F3. Each ORF is shown to scale as a box, numbered with the gene number. Each stop codon is represented as a green vertical line. The positions of the ΦM1 escape phage mutations are shown by red vertical lines, labeled with the parent phage. The scale is in base pairs.