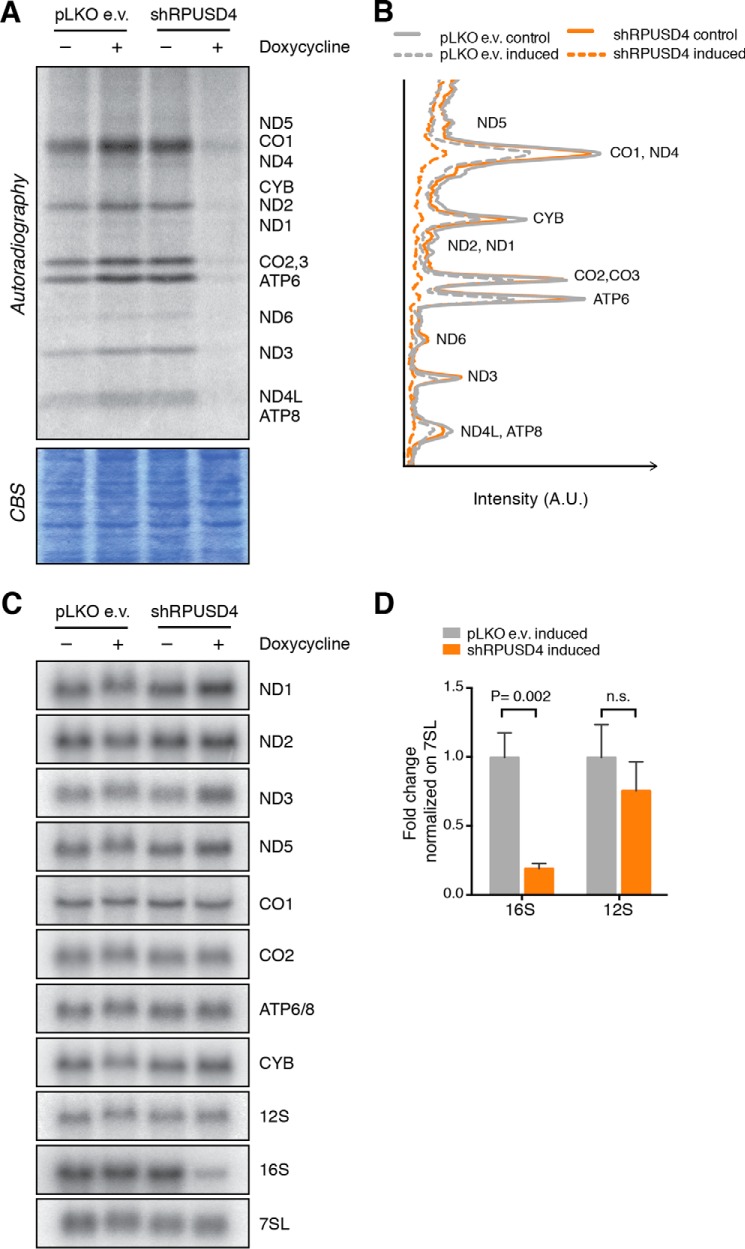
FIGURE 4.
Silencing of RPUSD4 affects mitochondrial protein translation by decreasing the 16S ribosomal RNA. A, 35S labeling of de novo mitochondrial protein synthesis in RPUSD4-silenced 143B cells with a region of the Coomassie Blue-stained gel (CBS) shown as a loading control. B, phosphorimaging quantification profile of the bands shown in A. C, Northern blotting analysis of total RNA extracts from RPUSD4-silenced 143B cells (shRPUSD4) and control cells (pLKO e.v.) probed for the RNA sequences as indicated. Cytosolic 7SL RNA was used as a loading control. D, densitometric quantification of the results for the rRNAs shown in C. The values for 16S and 12S rRNAs were normalized to the value obtained for the 7SL RNA and expressed as the -fold change with respect to the value obtained for pLKO e.v. cells treated with doxycycline. The combined data from four independent experiments are shown as means; error bars represent S.D. p values were obtained using Student's t test. CYB, cytochrome b.