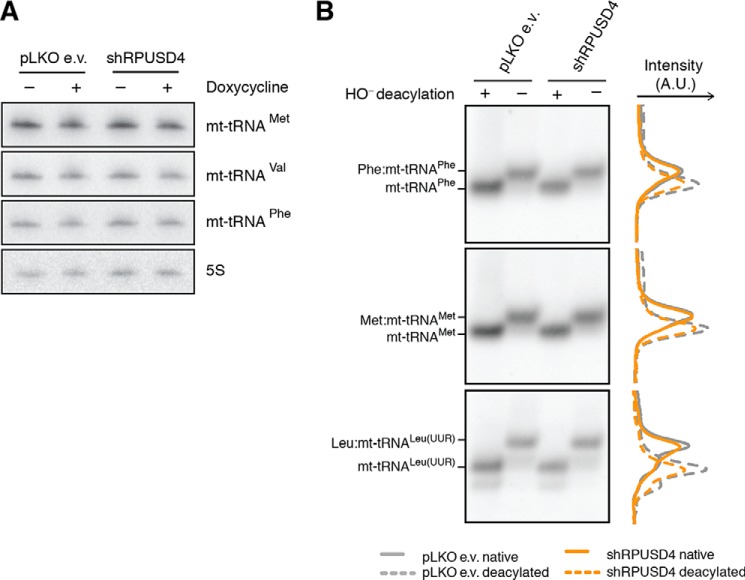
FIGURE 7.
Defective pseudouridylation of mt-tRNAPhe does not affect its stability or its aminoacylation. A, high resolution Northern blotting analysis of total RNA extracts from RPUSD4-silenced 143B cells (shRPUSD4) and control cells (pLKO e.v.) probed for mt-tRNAMet, mt-tRNAVal, and mt-tRNAPhe. Cytosolic 5S rRNA was used as a loading control. B, analysis of aminoacyl mt-tRNAs by high resolution Northern blotting of total RNA from pLKO e.v.- or shRPUSD4-expressing cells isolated under acid conditions. Where indicated the RNA was treated with alkali (OH−) to deacylate tRNAs. Membranes were probed for mt-tRNAPhe, mt-tRNAMet, and mt-tRNALeu(UUR). Densitometry profiles are presented for each mt-tRNA on the right side of the respective blot. A.U., arbitrary units.