FIGURE 2.
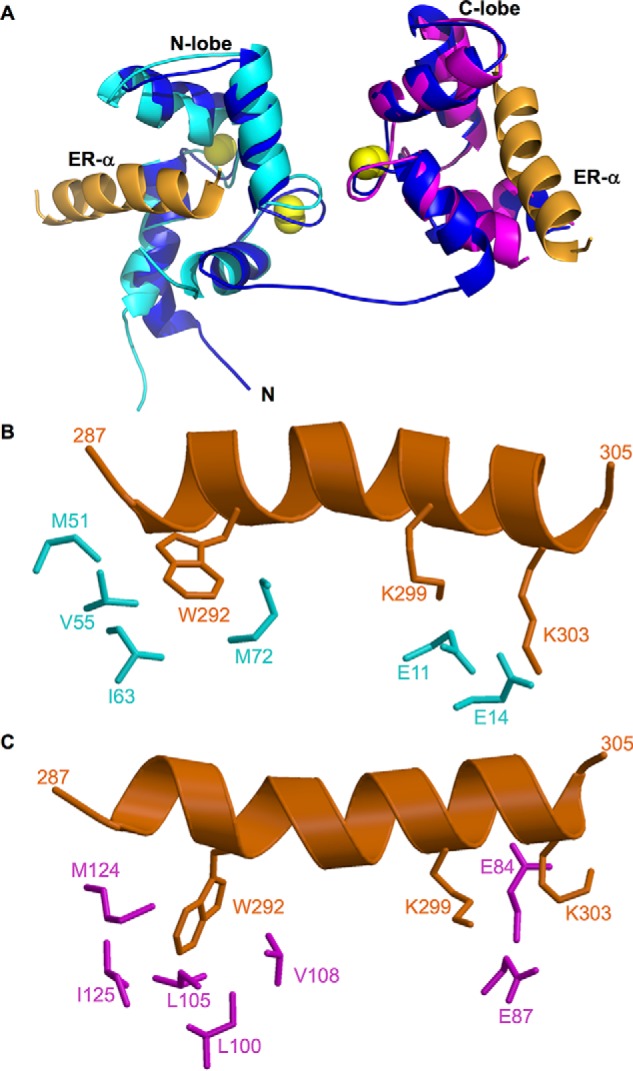
Structures of CaM·ER-α complexes. A, NMR structure of full-length CaM (dark blue) bound to two molecules of ER(287–305) (orange). The atomic coordinates were deposited into the Protein Data Bank (PDB accession no. 5T0X). Previous NMR structures of CaM-N (2LLO, cyan) and CaM-C (2LLQ, magenta) are overlaid on the full-length CaM structure for comparison. Bound Ca2+ are yellow spheres. The flexible interdomain linker (residues 74–82) forms an extended main chain conformation depicted by a blue line. B and C, exposed hydrophobic side chain atoms of CaM N-lobe (highlighted cyan in panel B) and C-lobe (highlighted magenta in panel C) interact with side chain atoms of key ER-α residues (orange) shown as sticks. Hydrophobic side chain atoms in ER-α (Trp-292) form detailed contacts with each lobe of CaM, and basic side chains in ER-α (Lys-299 and Lys-303) form salt bridges with Glu-14 (N-lobe) and Glu-84 (C-lobe) of CaM.
