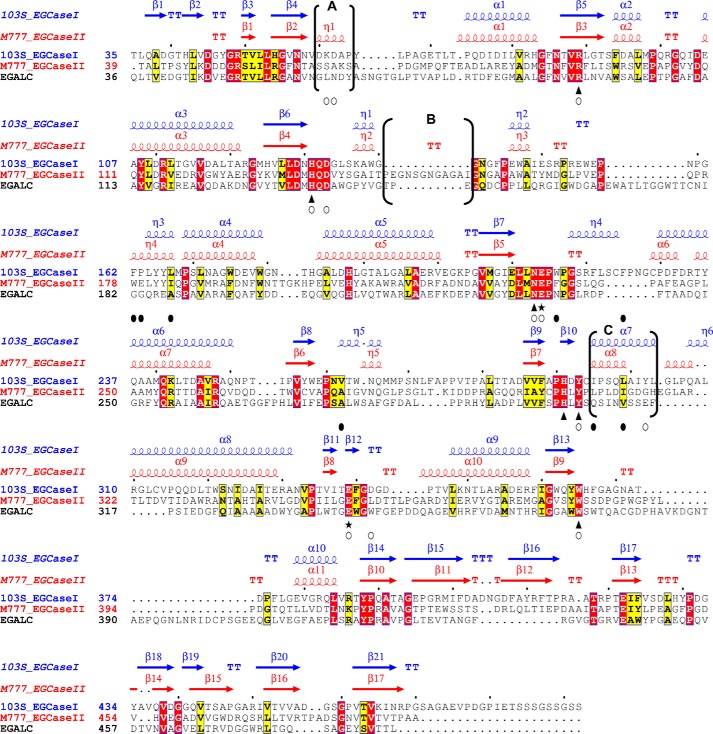
FIGURE 7.
Structure-based sequence alignment of 103S_EGCase I, M777_EGCase II and EGALC. The amino acid sequences of 103S_EGCase I (GenBankTM accession number CBH49814), M777_EGCase II (GenBankTM accession number AAB67050), and EGALC (GenBankTM accession number BAF56440) were aligned using PROMALS3D and shaded in ESPript 3.0. Identical residues are shown in open boxes with white letters on a red background. Similar residues are shown in open boxes with black letters on a yellow background. Conserved amino acid residues in the GH5 family of glycosidases are indicated by triangles. Two glutamates, functioning as an acid/base catalyst and nucleophile, respectively, are indicated by stars. Residues that form hydrogen bonds or hydrophobic interactions with the sugar moiety are indicated by empty circles. Residues that form the hydrophobic tunnel are indicated by black filled circles. The secondary structural elements are shown above the amino acid residues in blue (103S_EGCase I, PDB code 5J7Z) and red (M777_EGCase II, PDB code 2OSX). The major differences in the secondary structures of 103S_EGCase I and M777_II are indicated in brackets and marked as regions A, B, and C.