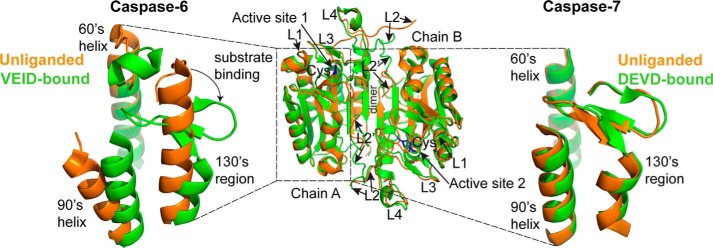
FIGURE 1.
Caspase-6 undergoes helix-strand transition upon substrate binding. The overall fold of canonical caspases before and after substrate binding is represented by the superimposition of the unliganded (orange; PDB code 1K86) and the peptide-based substrate mimic DEVD-bound (green; PDB code 1F1J) structures of caspase-7 (middle). Highlighted regions are the active site cysteine (blue), the dimer interface, and the substrate binding loops 1–4 (L1–L4). Caspase-7, like all other caspases, adopts a canonical strand conformation in its 130's region in both the unliganded state (orange; PDB code 1K86) and the peptide-based substrate mimic DEVD-bound (green; PDB code 1F1J) states (right). In contrast, caspase-6 can adopt a noncanonical extended helical conformation in its 130's region in the unliganded state (orange, PDB code 2WDP) but recovers the canonical strand conformation upon binding to a peptide-based substrate mimic VEID-aldehyde (green; PDB code 3OD5) (left).