FIGURE 4.
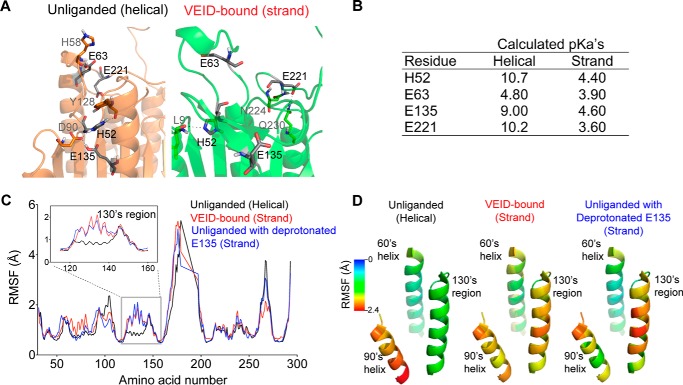
Protonation state of Glu-135 is critical to the dynamics of the 130's region. A, the unliganded (orange) and VEID-bound (green) states of caspase-6 present different local charge states of four identified residues in close proximity and within the 130's region, which are deprotonated upon substrate binding (shown in black). B, calculated local pKa values of the four amino acid residues that undergo differential protonation states between the unliganded (helical) and the VEID-bound (strand) states of caspase-6. C, plot of RMSF (Å) of the backbone atoms of each amino acid in caspase-6. A 1.2-ns-long simulated annealing-based molecular dynamics simulation was performed in all capase-6 protonation variants. The unliganded helical protonation variant represents the control ensemble of the triply protonated state (HisH-52, GluH-135, GluH-221) of caspase-6. The VEID-bound (strand) protonation variant represents the control ensemble of the triply deprotonated state (His-52, Glu-135, Glu-221) of caspase-6. Unliganded with deprotonated E135, protonation variant of caspase-6 that has deprotonated Glu-135 but remains protonated at the other two residues (HisH-52 and GluH-221). Residue-by-residue fluctuations are shown for caspase-6 in the unliganded (helical) state (black), VEID-bound (strand) state (red), and the unliganded caspase-6 with deprotonated Glu-135 (blue). The RMSF profile of unliganded caspase-6 with deprotonated Glu-135 behaves similarly to the VEID-bound (strand) state in the 130's region (inset). D, variation of the RMSF (Å) along key regions of caspase-6 mapped onto the structure of unliganded caspase-6 (PDB code 2WDP) highlighting only the 60's, 90's, and 130's regions.