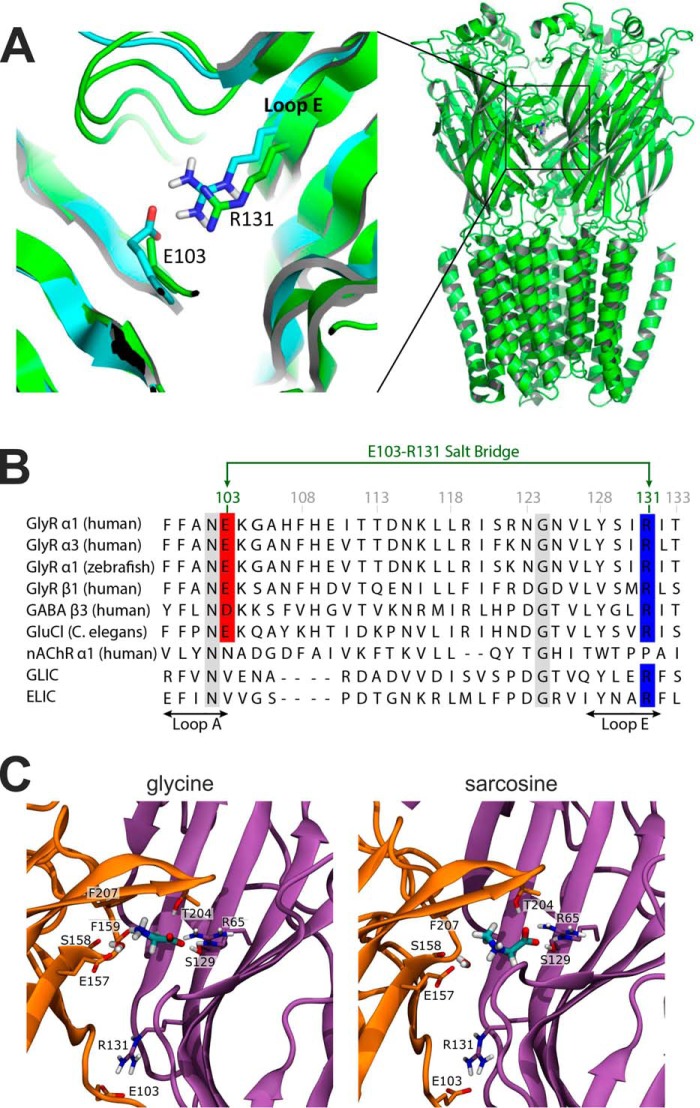
FIGURE 1.
Homology modeling reveals a salt-bridge between residues Glu-103 and Arg-131. A, location of the Glu103-Arg131 salt-bridge as predicted by a GlyR homology model (cyan)(7) built using GluCl as template (PDB code 2XYS) (16). The model is overlaid with the recent cryo-EM structure (green) of the zebrafish α1 GlyR (PDB 3JAE) (17). Note that the Glu-103 side chain is not fully resolved in the cryo-EM structure, but is certainly within distance to form the salt bridge as postulated by the model. B, an amino acid alignment of this region for nine members of the pLGIC superfamily showing the position of the salt bridge between Glu-103 (red) and Arg-131 (blue) and its conservation in glycine receptors. Other conserved positions are indicated by a gray background. C, the agonist binding mode and the Glu103-Arg131 salt bridge in a GlyR homology model based on the zebrafish α1 GlyR structure (PDB 3JAE) (17). Glycine (left) and sarcosine (right) are shown in the final frame after 500 ns simulation time. Orange, principal subunit; purple, complementary subunit. For both ligands, the carboxyl group is sandwiched by hydrogen bonds with Thr-204 and Ser-129 and is further stabilized by a salt bridge with Arg-65. The ammonium moiety interacts with a water molecule in the binding pocket, and the water is stabilized by hydrogen bonds to Glu-157 and to the backbone carbonyl oxygen of Ser-158 (as in Ref. 7). The ammonium group of glycine engages in an additional cation-π interaction with Phe-207 and forms a hydrogen bond with the backbone carbonyl group of Phe-159. This is sterically hindered for sarcosine by its additional N-methyl group.