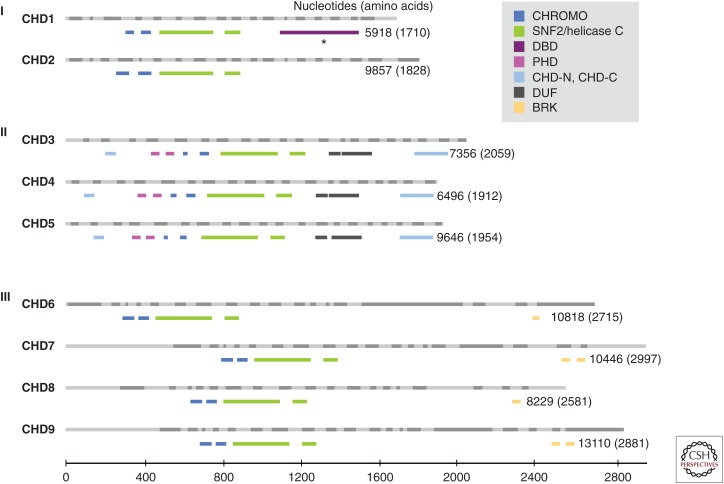
Figure 1.
The chromodomain helicase DNA-binding (CHD) family of chromatin remodelers. CHD proteins are classified into three subfamilies (Roman numerals) based on their functional motifs (see legend). The human CHD family based on Ensembl is drawn to scale, with light and dark gray bars depicting alternating exons (above) and the functional motifs from PFAM (a database of protein families of multiple sequence alignments generated using hidden Markov models) shown in color (below) for each CHD member. The number of nucleotides and amino acid residues for the CHD transcript and protein, respectively, are shown. BRK, Brahma and Kismet domains; CHD, chromodomain helicase DNA binding; CHROMO, chromodomain; CHD-N, CHD-C: CHD_N and CHD_C are shown in upstream and downstream regions, respectively; DBD, DNA-binding domain (based on Delmas et al. 1993) rather than PFAM; DUF, domain of unknown function; PHD, plant homeodomain; SNF2/Helicase C, SNF2_N, and Helicase_C are shown in upstream and downstream regions, respectively. (From Li and Mills 2014; reproduced and modified, with express permission, from Future Medicine, 2014.)