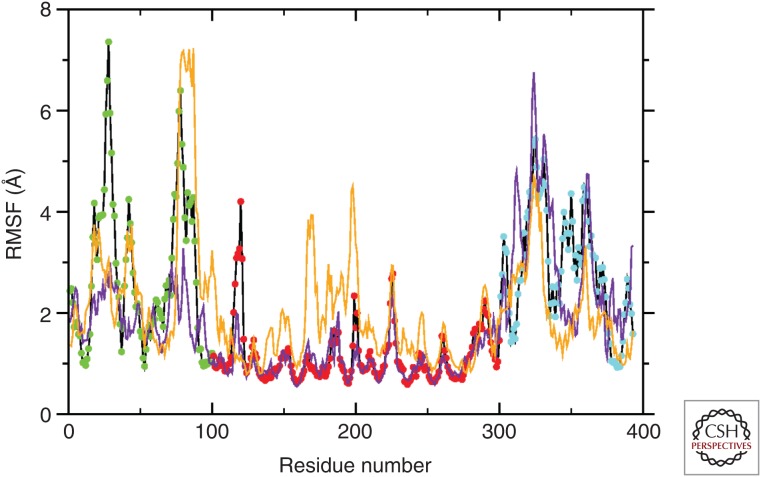
Figure 4.
Per-residue root-mean-square fluctuation (RMSF) for wild-type p53 and the R175H mutant. The average fluctuation observed by molecular dynamics (MD) (200 nsec) shows different degrees of movement depending on the distinct region of p53 in the wild-type form (black line); point color code as in Fig. 1 (amino terminal, green; DBD [DNA-binding domain], red; carboxyl terminal, blue). The two TADs (transactivation domains), PRR (proline-rich domain), OD (oligomerization domain), and CTD (carboxy-terminal domain) are clearly distinguishable. The DBD is highly stable. Mutant p53 isoforms shows a drastic change in flexibility. The p53 R175H mutant that maintains the native Zn binding (yellow line) has increased the flexibility of DBD, indicating a potential abnormal binding to the selected promoter. The p53 R175H mutant that has an alternative Zn-binding site (purple line) has reduced flexibility of the carboxyl terminus, suggesting abnormal transactivation properties. (From D’Abramo et al. 2015; modified, with permission.)