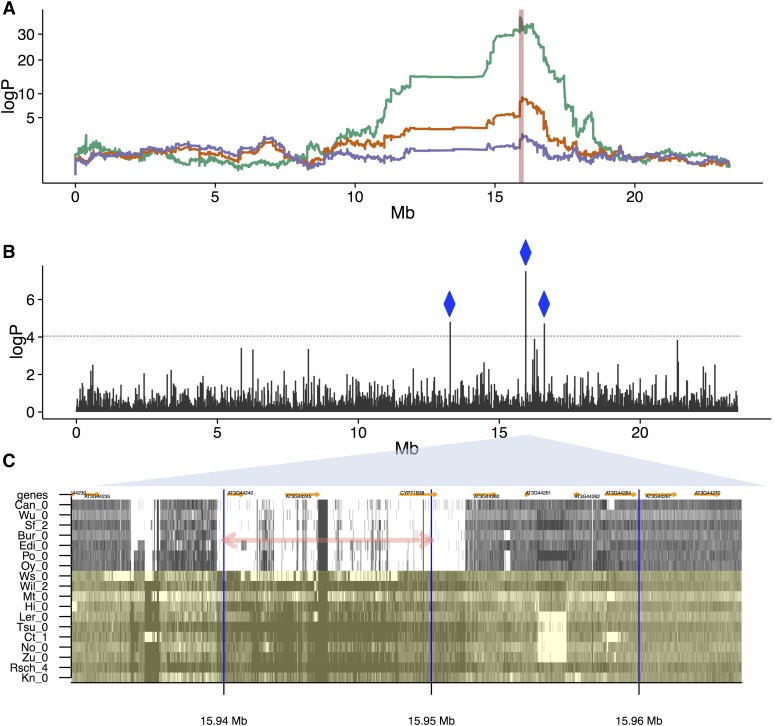
Figure 5.
Effects of SVs on germination time. (A) Genome scans over chromosome 3 (x-axis: genomic position, y-axis: logP of association). Orange: association of local haplotype with germination time (days), peaking at 15.93 Mb. Green: association of local haplotype with the SV trait unpaired reads at the source locus 15.94–15.95 Mb (indicated by the vertical red line), explaining 8.13% of the variance in germination time, with an SV-QTL mapped at the same position as the germination QTL. Purple: residuals of germination time after regressing out the SV trait, ablating the QTL. (B) Chromosome-wide Pearson correlations between germination time and the numbers of unpaired reads measured at each 10 kb source locus (x-axis: genomic position, y-axis: −log10 P-value of test that the correlation is zero). Three source loci correlate strongly with germination (logP > 4), all with cis SV-QTL (blue diamonds). (C) Structural variation in the MAGIC founders. Shown is the read coverage in 18 accessions (labeled on y-axis),covering ∼30 kb surrounding ∼15.94 Mb (x-axis). Dark shades indicate high coverage, light shades low coverage. The 10 kb intervals used to define source loci are delineated by vertical blue lines. The source locus giving rise to the SV-QTL in (A), (B) is marked with a pink double-arrow. Those founder accessions predicted to carry the reference allele (No-0, Ct-1, Mt-0, Wil-2, Ler-0, Tsu-0, Rsch-4, Kn-0, Zu-0, Hi-0, and Ws-0) are in green, those predicted to carry the SV are in gray. Genes are annotated in orange.