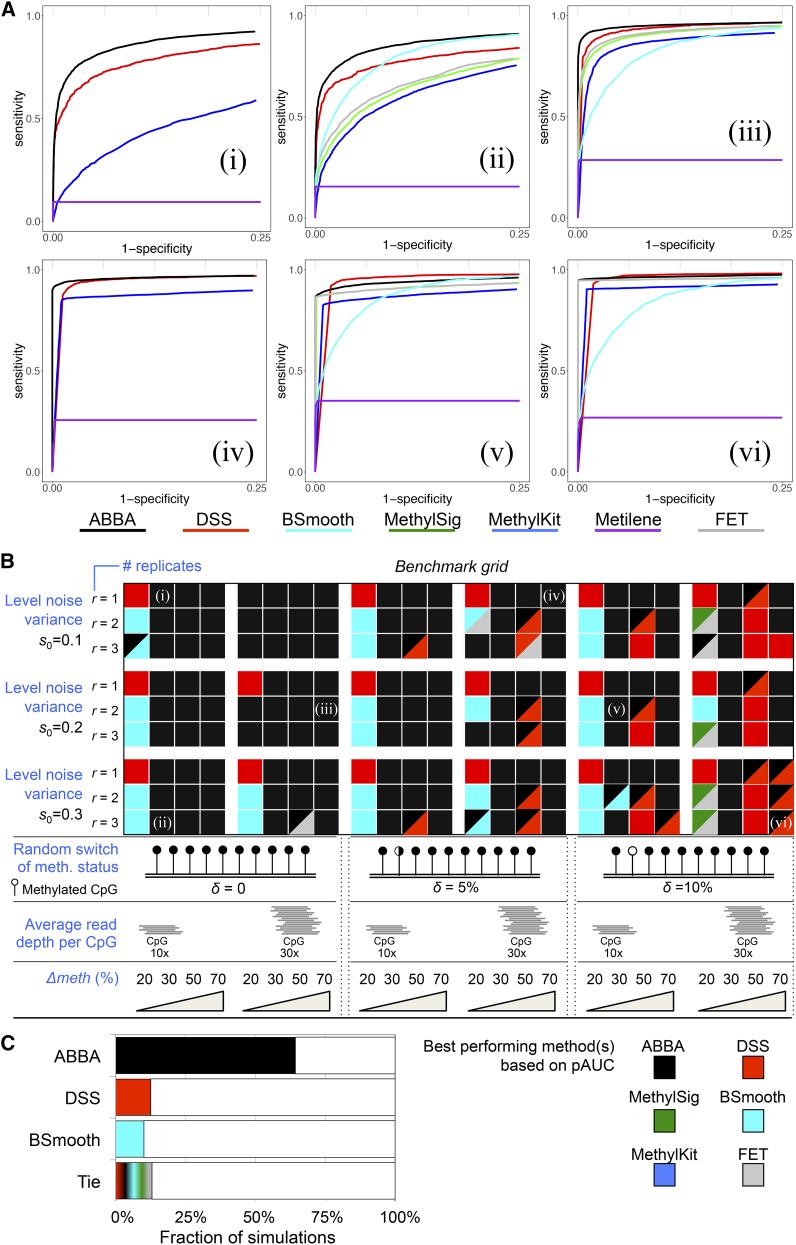
Figure 2.
Benchmarking results. (A) ROC curves for selected combinations of parameters: (i) s0 = 0.1, Δmeth = 30%, r = 1, average read depth per CpG of 10×, δ = 0; (ii) s0 = 0.3, Δmeth = 30%, r = 3, average read depth per CpG of 10×, δ = 0; (iii) s0 = 0.2, Δmeth = 70%, r = 2, average read depth per CpG of 30×, δ = 0; (iv) s0 = 0.1, Δmeth = 70%, r = 1, average read depth per CpG of 30×, δ = 5%; (v) s0 = 0.2, Δmeth = 30%, r = 2, average read depth per CpG of 10×, δ = 10%; (vi) s0 = 0.3, Δmeth = 70%, r = 3, average read depth per CpG of 30×, δ = 10%. For each of this combination of parameters, the corresponding best method based on its pAUC is indicated in the benchmark grid below. In (i) and (iv), ROC curves are reported only for the methods that can analyze WGBS data generated from one biological sample. (B) Global snapshot of the method’s performance across 216 simulated datasets. A given combination of parameters is indicated by a square in the benchmark grid, and, for each square, we calculated the pAUC for each method and determined which method had the overall best pAUC (i.e., pAUCmethod_1 > pAUCmethod_2). Colors in the benchmark grid indicate which method had the best performance. When pAUC of two methods are similar (±1%) we report the colors of both methods (e.g., black and red colors in the same square indicate similar performance of ABBA and DSS). The six selected combination of parameters for which the ROC curves are reported in (A) are indicated within the benchmark grid: (i–vi). All ROC curves are reported in Figure S5, Figure S6, and Figure S7. (C) For the three best performing methods (ABBA, DSS, and BSmooth), we report the percentage of simulated scenarios in which each method resulted to be the best based on the pAUC comparison. “Tie” indicates the proportion of simulated scenarios in which the pAUCs of any two methods were similar (i.e., pAUCs ±1%), and it was not possible to single out a single best performing approach.