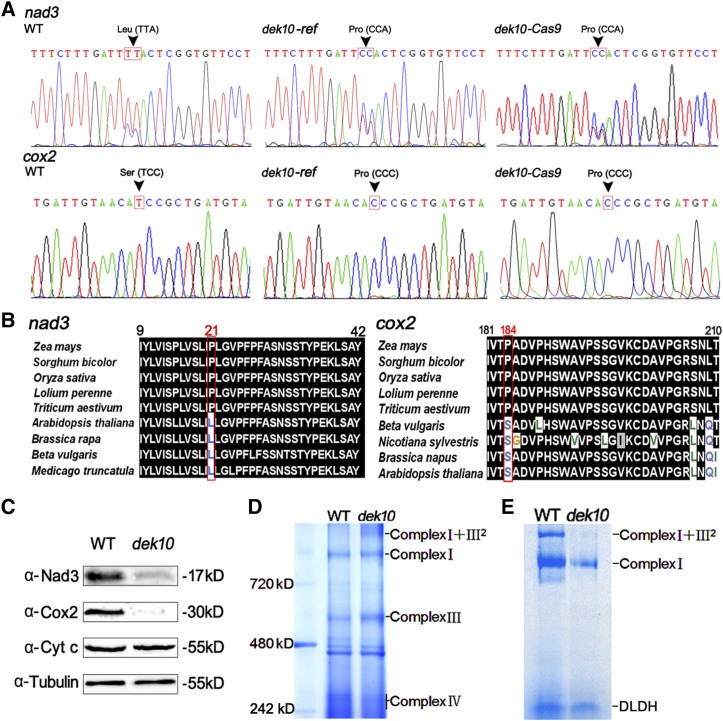
Figure 5.
Dek10 is required for nad3-61, nad3-62, and cox2-550 editing in maize mitochondria. (A) Analysis of RNA editing at the nad3-61, nad3-62, and cox2-550 sites in the transcripts from developing kernels of the WT, dek10-ref, and dek0-Cas9 mutants at 18 DAP. The arrow marks the editing site. (B) Alignment of the Nad3 sequences around amino acid 21 and alignment of the Cox2 sequences around amino acid 184. The protein sequences are derived from Z. mays, Sorghum bicolor, O. sativa, Lolium perenne, T. aestivum, Beta vulgaris, Nicotiana sylvestris, B. napus, M. truncatula, and Arabidopsis thaliana. Numbers indicate amino acid positions in the protein. The red box indicates the position affected by the nad3-61, nad3-62, and cox2-550 editing. (C) Western blot analysis with antibodies against Nad3, Cox2, and Cyt-c. Anti-Tub was used as sample loading control. (D) BN-PAGE of mitochondrial complexes. The positions of super complex I+III2, complex I, complex III, and complex IV are indicated. (E) In-gel NADH dehydrogenase activity test analysis of complex I activity. The positions of super complex I+III2 and complex I are indicated. The activity of the dehydrolipoamide dehydrogenase was used as a sample loading control. BN-PAGE, blue native polyacrylamide gel electrophoresis; Cox, Cyt-c oxidase; Cyt-c, cytochrome c; DAP, days after pollination; WT, wild type.